Figure 5.
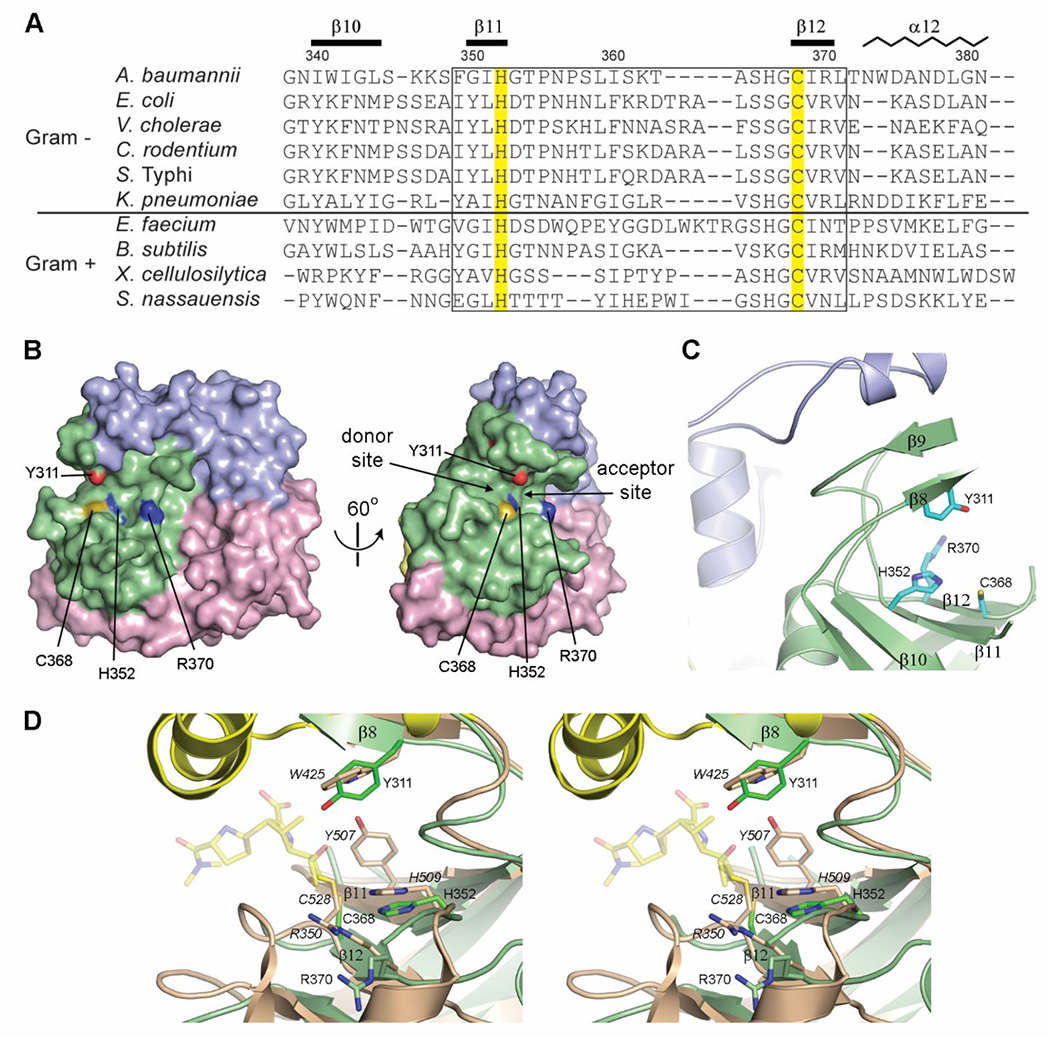
The LdtAb active site. (A) Partial structure-based sequence alignment of the active site of known Gram-positive and Gram-negative LDTs. The two catalytic residues are highlighted yellow. The secondary structure and residue numbering for LdtAb are shown above the sequences. The fingerprint LDT catalytic motif is indicated by the box. (B) Surface representation of LdtAb (left) and the same representation rotated by 60° on the right. The two catalytic residues (His352 and Cys368) delineate the donor and acceptor regions of the active site. (C) The LdtAb active site. (D) Stereoview of the LdtAb active site (light green ribbons and sticks) superimposed onto the E. coli YcbB LDT-ertapenem acyl-enzyme complex (light brown ribbons and sticks, with ertapenem shown in semitransparent yellow sticks, PDB ID 6NTW). The catalytic residues are indicated (E. coli labels are given in italics) along with the conserved Arg370 (Arg350 in E. coli). A tyrosine residue thought to be involved in protonation of the leaving groups in the E. coli enzyme (Tyr507) is spatially equivalent to a glycine (Gly350) on strand β11 (not shown for clarity) in LdtAb. A tyrosine residue (Tyr311) in LdtAb, in the equivalent position as a tryptophan in E. coli (Trp425), is near the catalytic residues and could play the same role postulated for Tyr507 in E. coli.
