Figure 6.
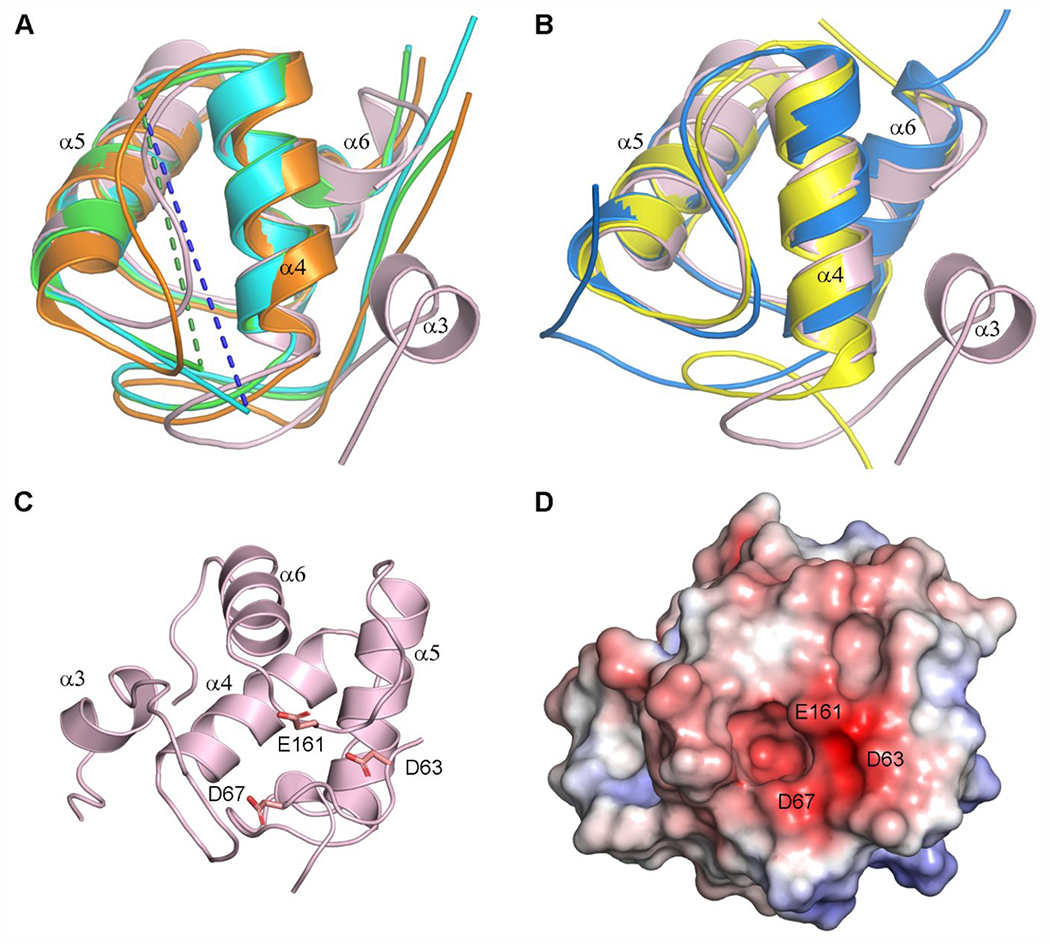
Structural comparisons of the LdtAb PG domain. (A) Superposition of the LdtAb PG domain (pink ribbons) against the equivalent domains in the LDTs from E. coli (green, PDB ID 6NTW), C. rodentium (cyan, PDB ID 7KGM), and V. cholerae (orange, PDB ID 7AJ9). The helices are labeled according to LdtAb secondary structure nomenclature. Missing α4–α5 loops in the E. coli and C. rodentium enzymes are indicated by green and blue dashed lines. (B) Superposition of the LdtAb PG domain (pink ribbons) against the equivalent domains in the LDTs from X. cellulosilytica (yellow, PDB ID 4LPQ) and S. nassauensis (blue, PDB ID 5BMQ). The helices are labeled according to LdtAb secondary structure nomenclature. (C) Ribbon representation of the LdtAb PG domain (pink) showing three clustered acidic residues. (D) Electrostatic surface representation of the LdtAb PG domain in the same orientation as panel C. A negatively charged surface patch resulting from the clustering of the three acidic residue is indicated.
