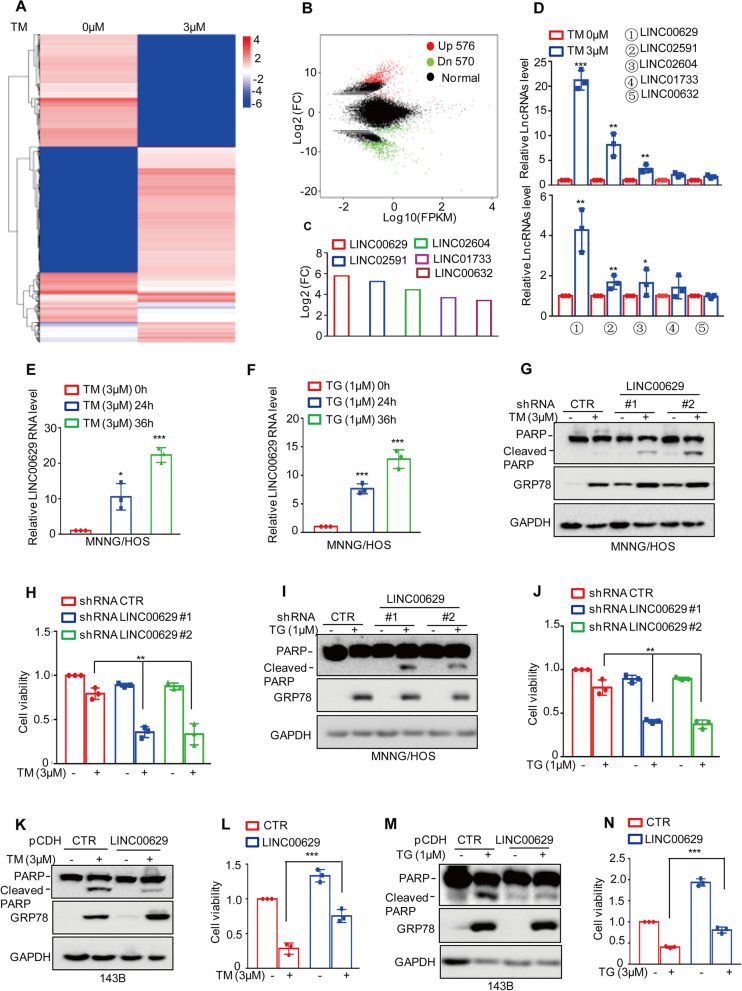
Fig. 1.
LINC00629 was increased by ER stress and suppressed cell apoptosis (A-B) MNNG/HOS cells were treated with 3 μM TM for 36 h. The altered LncRNAs were investigated by RNA sequencing analysis. C The significantly altered LncRNAs are listed. D MNNG/HOS and 143B cells were treated with 3 μM TM for 36 h. The expression levels of the LncRNAs were measured by qRT–PCR in MNNG/HOS (up) and 143B cells (down). E MNNG/HOS cells were treated with 3 μM TM for the indicated times. The expression levels of LINC00629 were measured by qRT–PCR. F MNNG/HOS cells were treated with 1 μM TG for the indicated times. The expression levels of LINC00629 were measured by qRT–PCR. G-H MNNG/HOS cells with or without LINC00629 knockdown were treated with or without 3 μM TM for 36 h. Cell apoptosis was detected by Western blot (G), and cell viability was analysed by CCK8 assay (H). GRP78 was used as the ER stress marker, and GAPDH was used as the loading control. I-J MNNG/HOS cells with or without LINC00629 knockdown were treated with or without 1 μM TG for 36 h. Cell apoptosis was detected by Western blot (I), and cell viability was analysed by CCK8 assay (J). (K-L) LINC00629 was overexpressed in 143B cells using lentivirus expressing the pCDH vector, and then the cells were treated with or without 3 μM TM for 48 h. Cell apoptosis was detected by Western blot (K), and cell viability was analysed by CCK8 assay (L). M-N 143B cells with or without LINC00629 overexpression were treated with 1 μM TG for 48 h. Cell apoptosis and cell viability were detected by Western blot (M) and CCK8 assays (N). Data in D, E, F, H, J, L, N were analysed by Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001