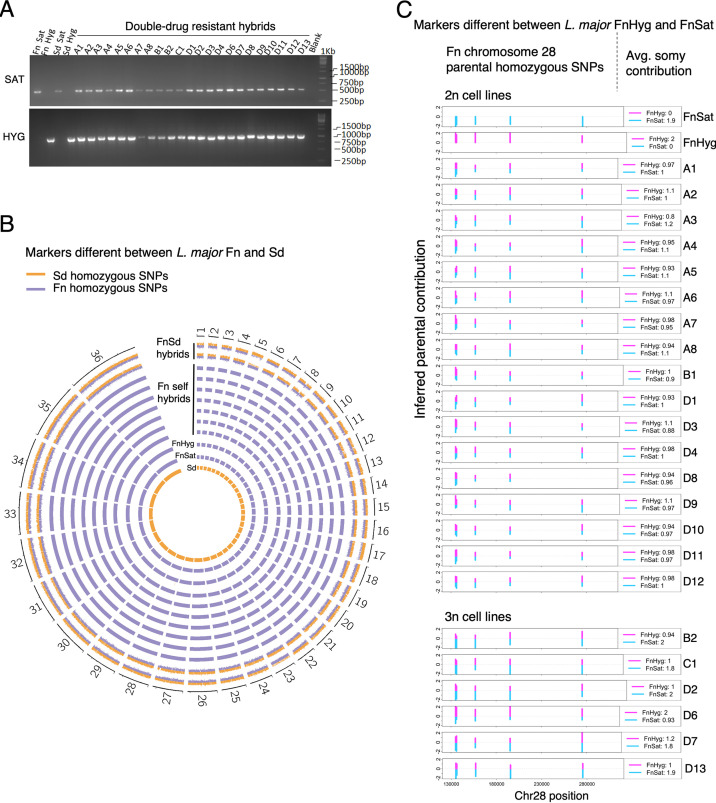
FIG 2.
Genotyping of selfing hybrids. (A) PCR confirmation of the selectable parental markers Sat and Hyg in parental and hybrid progeny clones. Selfing hybrids shown were generated from crosses between FnSat and FnHyg in the presence or absence of LmSd to evaluate the impact of including a heterologous strain on the selfing frequencies; 1 kb: DNA ladder. (B) Circos plots of the frequencies of parental SNP markers that are homozygous and distinct between Sd (orange bars) and Fn lines (blue bars). Genome-wide heterozygosity of the parental markers was observed in the FnSd hybrids generated previously (12) (FnSd1 and FnSd2). Only Fn markers were detected in the DDR hybrids, allowing us to conclude that they are selfing hybrids. Data from seven representative selfing hybrids are shown (A1, A2, A3, A4, C1, D1, and D7). The frequencies of the parental alleles are represented as histogram bars for each genome position. Chromosome labels are shown on the outer circle. (C) Bottlebrush plots showing biparental inheritance of homozygous SNPs that are different between FnHyg and FnSat on chromosome 28 (chr28). Blue and magenta bars indicate SNPs mapping to the FnSat and FnHyg parents, respectively. The vertical distances correspond to the inferred allelic depth that was normalized across the genome which was assigned an average ploidy of 2n or 3n (selfing hybrids B2, C1, D2, D6, D7 and D13). The average somy contribution of the respective Fn parents is shown on the right inset of each plot. The data for one of the allelic markers on self-hybrid D8 was lost due to filtering.