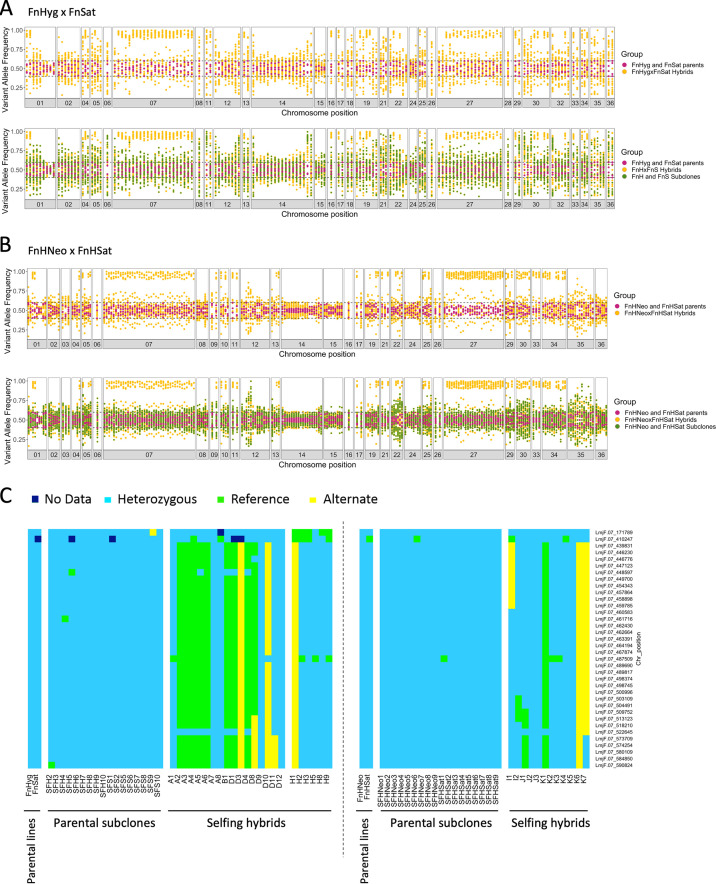
FIG 3.
Loss of heterozygosity in selfing hybrids. Segregation pattern of SNPs that are heterozygous in both parents is shown by the frequency of variant alleles identified on different chromosomes for 17 FnHyg x FnSat 2n hybrids (A) and 12 FnHNeo × FnHSat 2n hybrids (B). Parental SNP frequencies are shown in magenta, the corresponding allele on the same position are shown as yellow dots for the 2n hybrid progeny. The corresponding allele frequencies of the 2n parental subclones are represented by green dots in the bottom panels; 9 subclones of FnHyg and 8 subclones of FnSat in A, 9 subclones of FnHNeo and 9 subclones FnHSat in B. SNPs were considered heterozygous if their frequencies were between 0.4 and 0.6 on that genomic position, and homozygous if ≥ 0.85. Allele frequencies of 1.0 were subtracted by a random number between 0 and 0.05 before plotting to transform them into frequencies of 1.0 to 0.95 for a clearer visualization of data points. (C) Inheritance of parental heterozygous SNPs in high SNP density locus on chromosome 7 (position 439831 to 590824). Individual SNPs are indicated on the y axis on the right side of the panel and the type of SNP (heterozygous, homozygous matching L. major reference genome sequence and homozygous matching the alternate allele) is indicated by color. SFH and SFS refer to subclones derived from FnHyg and FnSat, respectively, and the 2n hybrid progeny are labeled A1 to H9. No significant difference between observed and expected frequencies: Chi square value, 0.7115; degrees of freedom, 2; P = 0.7007. SFHNeo and SFHSat refer to subclones derived from FnHNeo and FnHSat, respectively, and the 2n hybrid progeny are labeled I1 to K7. The segregation pattern of all homozygous and heterozygous SNPs in both parents is shown in Fig. S6.