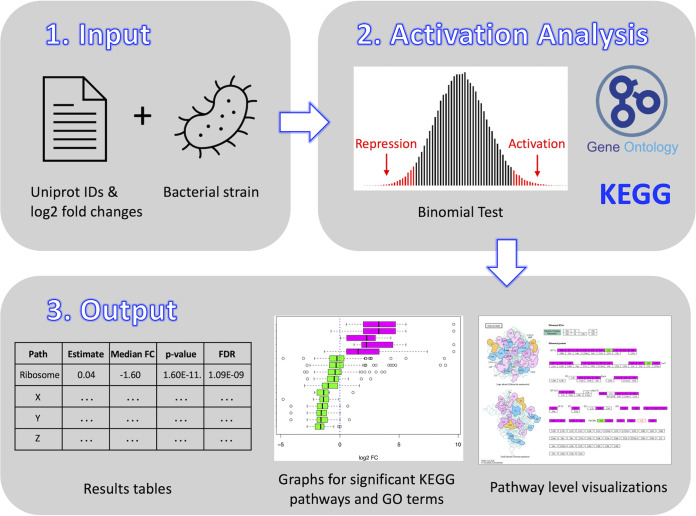
FIG 1.
ESKAPE Act PLUS workflow. An analysis starts with upload of a file containing Uniprot identifiers and log2 fold changes as well as selection of a bacterial strain matching the identifiers in the input file. ESKAPE Act PLUS uses these user-provided inputs as well as KEGG and Gene Ontology (GO) annotations to perform an activation analysis using a binomial test to identify significantly activated or repressed KEGG pathways or GO terms. Outputs include tables with results for all KEGG pathways and GO terms, graphs for significantly activated or repressed KEGG pathways and GO terms, and links to pathway level visualizations of induced and repressed genes or proteins for significant KEGG pathways generated with the KEGG Mapper Color Tool (14, 15). The Gene Ontology logo is available under a CC BY 4.0 license and the KEGG ribosome pathway image is used with permission.