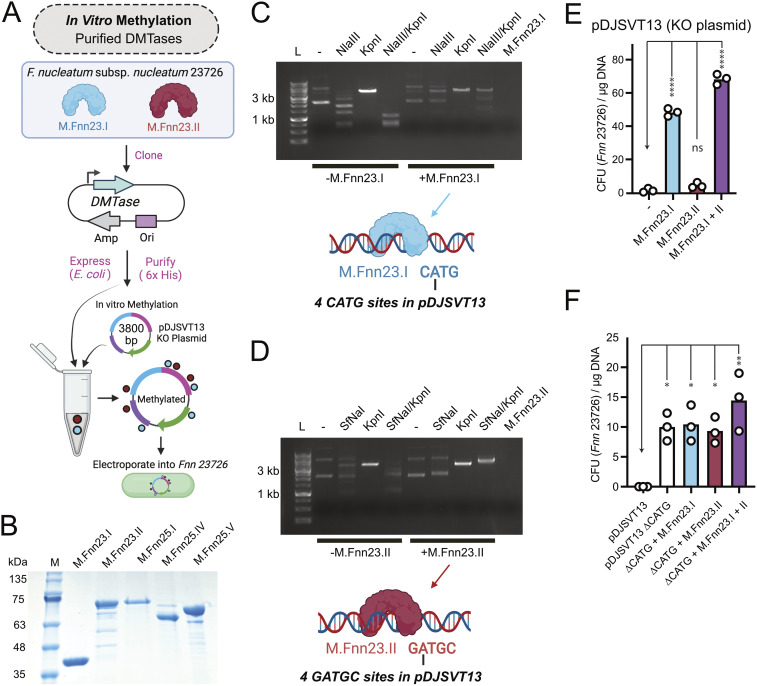
FIG 2.
F. nucleatum MTases protect plasmid DNA and allow for more efficient chromosomal plasmid incorporation in F. nucleatum subsp. nucleatum 23726. (A) Schematic of our process to produce recombinant MTases that were next used to treat plasmid DNA in vitro prior to electroporation into F. nucleatum subsp. nucleatum 23726. (B) SDS-PAGE gel of five purified MTases from F. nucleatum subsp. nucleatum 23726 and F. nucleatum subsp. nucleatum 25586. (C) Methylation of pDJSVT13 with M.Fnn23.I protects against DNA cleavage by the REase NlaIII (NEB), which cuts at CATG sites. KpnI was used as a unique single-cut enzyme in pDJSVT13. The M.Fnn23.I lane is enzyme alone and shows no contaminating DNA brought in with the pure protein. (D) Methylation of pDJSVT13 with M.Fnn23.II protected against DNA cleavage by the REase SfNaI (NEB), which cut at GATGC sites. (E) Methylation of pDJSVT13 resulted in an increased number of transformants (in CFU per microgram of DNA). (F) By changing the four CATG sequences to CACG (ΔCATG), which are the target for the MTase M.Fnn23.I, transformation efficiency was significantly increased, even in the absence of methylation. Statistical values are as follows: ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. For panels E and F data, we used a two-way ANOVA.