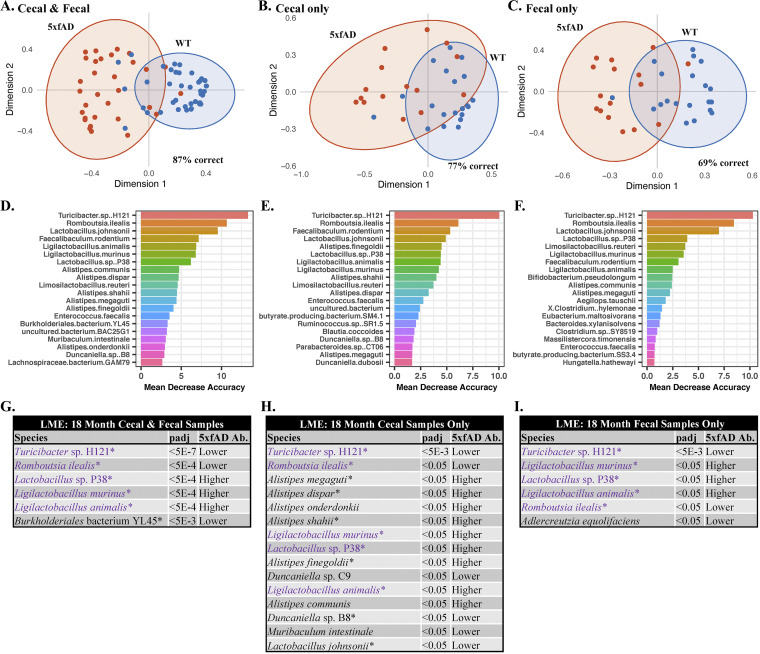
FIG 4.
Random forest and linear mixed effects (LME) modeling of microbiomes from 18-month-old animals. All evaluations were performed with separation by genotype (WT versus 5xfAD). Random forest (A–F) of the cecal and fecal samples combined (A and D), the cecal samples only (B and E), and the fecal samples only (C and F). Turicibacter sp. H121 was responsible for the largest proportion of separation in all of the random forest analyses, with a mean decreased accuracy from 10 to 14%. LME (G–I) was performed with Housing ID as the random variable. Shown are the cecal and fecal samples combined (G), the cecal samples only (H), and the fecal samples only (I). The “5xfAD Ab.” column in panels G–I indicates whether the species was found in relatively higher or lower abundance in the 5xfAD samples, compared to the WT samples. The species in purple were found to be significantly different in all three comparisons, and species with an asterisk were among the top drivers of separation in the random forest analysis. The LME computed P values (P adj) are false-discovery rate (FDR) corrected.