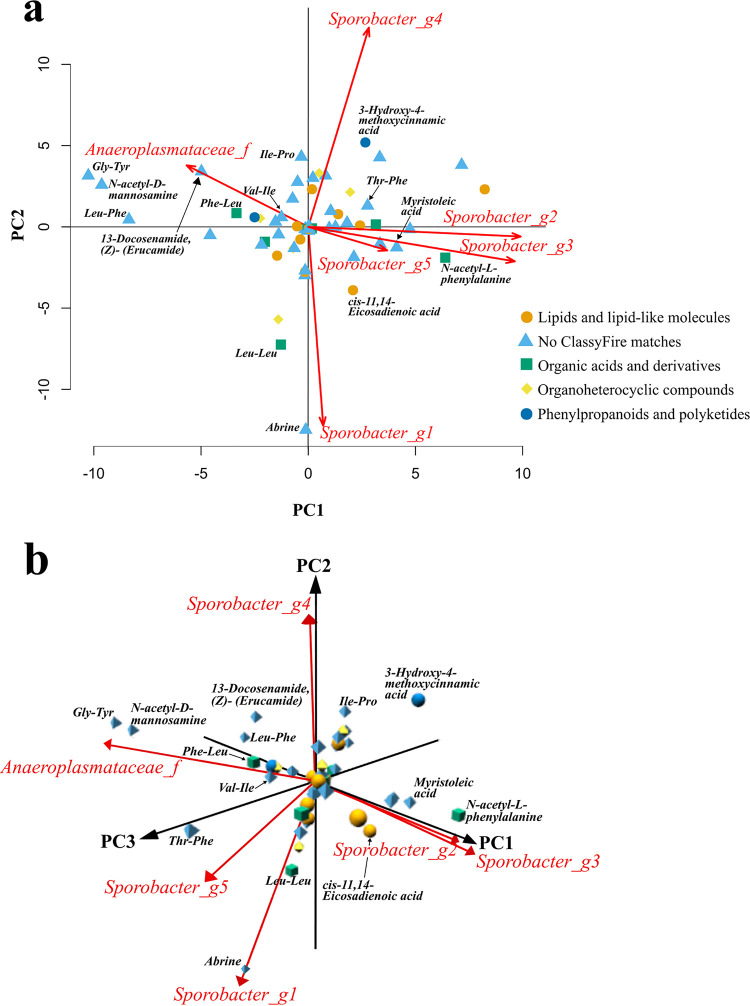
FIG 3.
Principal-component analyses (PCA) illustrate probable metabolite-microbe cooccurrences. Derived from analyses where n = 90 (Norman n = 18, Guayabo n = 12, Tambo de Mora n = 14, Boulkiemdé n = 11, Tunapuco n = 24, and Matses n = 11). Metabolite feature placements are based on conditional probabilities produced by mmvec (69). Annotated shared metabolite features from gap-filled data are represented as dots and are shape- and color-coded based on ClassyFire (87) assignments from MolNetEnhancer analyses (88). PCAs include biplots highlighting the most influential taxa across each principal component (PC) represented with red arrows showing their influence along the PCs. Taxonomic assignments were simplified to include unique identifiers for each label, such as “Anaeroplasmataceae_f” representing a read assigned to the family Anaeroplasmataceae. Multiple Sporobacter genera were identified and were given a “_g” label followed by a number for each instance of Sporobacter genera. (a) Two-dimensional representation of shared metabolite-microbe predicted interactions along PCs 1 to 2. Three taxa are represented for each component. Legend from panel a also applies to panel b. (b) Three-dimensional figure of shared metabolite-microbe predicted interactions across PCs 1 to 3. Two taxa are represented for each component.