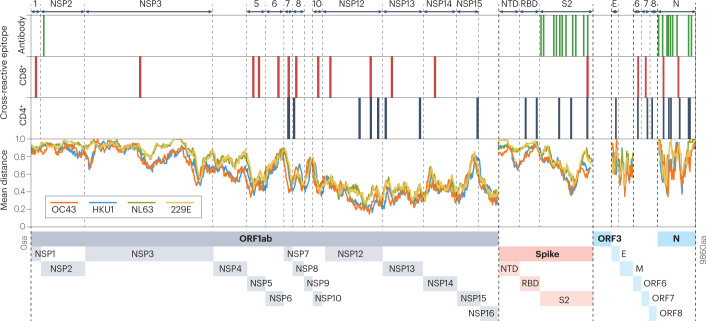
Fig. 2. Severe acute respiratory syndrome coronavirus 2 cross-reactive epitopes and common cold coronavirus homology.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteome with regions of experimentally defined cross-reactive epitopes in humans, as listed in Supplementary Table 1. SARS-CoV-2 proteins containing cross-reactive epitopes are labelled along the top and depicted along the bottom of the figure. The amino acid sequences for the ancestral SARS-CoV-2 and the reference sequences [Accession numbers: 229E: NC_002645.1, OC43: NC_006213.1, NL63: NC_005831.2, HKU1: NC_006577.2] for each of the four common cold coronaviruses were aligned against each other using MATLAB multialign function and the PAM250 scoring matrix. A moving window was used to calculate the mean number of amino acid (aa) differences between the ancestral SARS-CoV-2 sequence and each common cold coronavirus (mean number of mismatches per base (mean distance)). A window size of 100 was used for ORF1ab and spike proteins, and a window size of 30 was used for other proteins. E, envelope protein; M, membrane protein; N, nucleocapsid protein; NSP, non-structural protein; NTD, N-terminal domain; ORF, open reading frame; RBD, receptor-binding domain; S2, spike subunit 2.