FIG 1.
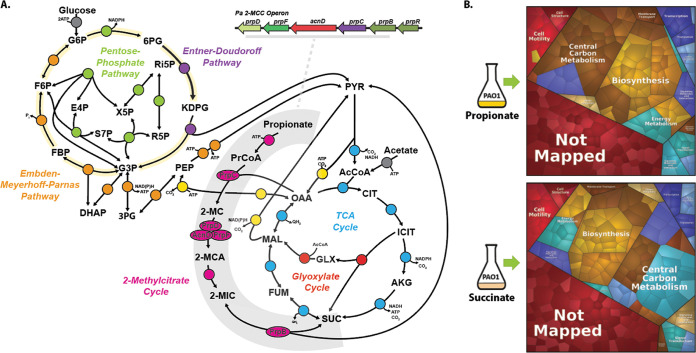
Proteomic analysis of Pa grown on succinate and propionate. (A) Schematic depicting the Pa 2-methylcitrate cycle (2MCC) in Pa central carbon metabolism. The Pa central metabolic network shown here consists of six main blocks, designated with different colors: (i) the Embden-Meyerhoff-Parnas pathway (EMP; orange); (ii) the pentose phosphate pathway (PPP; green); (iii) the Entner-Doudoroff pathway (EDP; purple); (iv) the tricarboxylic acid cycle (TCA; blue) and glyoxylate shunt (red); (v) anaplerotic and gluconeogenic reactions (yellow); and (vi) the 2MCC (pink). The 2MCC operon arrangement (inset, gray underline) consists of genes that encode a transcriptional regulator (designated here as prpR), which is thought to encode a ligand-responsive repressor, a methylcitrate synthase (prpC), which condenses propionyl-CoA (PrCoA) with oxaloacetate (OAA) to form 2-methylcitrate (2-MC), a 2-methylcitrate dehydratase/hydratase (prpD), which dehydrates 2-MC to yield 2-methylaconitate (2-MCA), a 2-methylcitrate dehydratase (acnD) and 2-methylaconitate cis-trans isomerase (prpF), which provide an alternative route for the generation of 2-MCA from 2-MC (the reason for an alternative route for 2-MCA generation in Pa is currently unclear), and a 2-methylisocitrate lyase (prpB), which cleaves 2-methylisocitrate (2-MIC) to yield pyruvate (PYR) and succinate (SUC). Note that the 2-MCA generated in the PrpD or AcnD/PrpF reactions is rehydrated by an unlinked aconitase (likely AcnB in Pa) to yield the PrpB substrate 2-MIC. Also, the enzyme responsible for the initial activation of propionate to yield PrCoA has not yet been identified for Pa, although in other organisms this function is carried out by a dedicated propionyl-CoA synthase (PrpE), by acetyl-CoA synthase (AcsA), by a combination of phosphotransacetylase (Pta) and acetate kinase (AckA) activities, or by an additional, uncharacterized propionyl-CoA ligase (7). AcCoA, acetyl-coenzyme A; CIT, citrate; ICIT, isocitrate; AKG, α-ketoglutarate; FUM, fumarate; MAL, malate; KDPG, 2-keto-3-deoxy-6-phosphogluconate; G3P, glyceraldehyde 3-phosphate; FBP, fructose 1,6-bisphosphate; F6P, fructose 6-phosphate; G6P, glucose 6-phosphate; 6PG, 6-phosphogluconate; Ri5P, ribulose 5-phosphate; R5P, ribose 5-phosphate; X5P, xylulose 5-phosphate; S7P, sedoheptulose 7-phosphate; E4P, erythrose 4-phosphate; PEP, phosphoenolpyruvate. (B) Illustration of the statistically significant proteomic changes (P ≤ 0.05, fold change of ≥1) during growth on propionate or succinate, as represented by Voronoi tessellations. Pathway assignment was performed using the KEGG data set. Proteome alterations that could not be assigned to a specific pathway (uncharacterised/hypothetical proteins) are shown as “Not Mapped.” The specific protein identities for the protein clusters that were upregulated during growth on propionate are shown in Fig. S1A in the supplemental material, and statistical analyses of these data are illustrated in Fig. S1B to D. The complete proteomics data set is presented in Data Set S1.