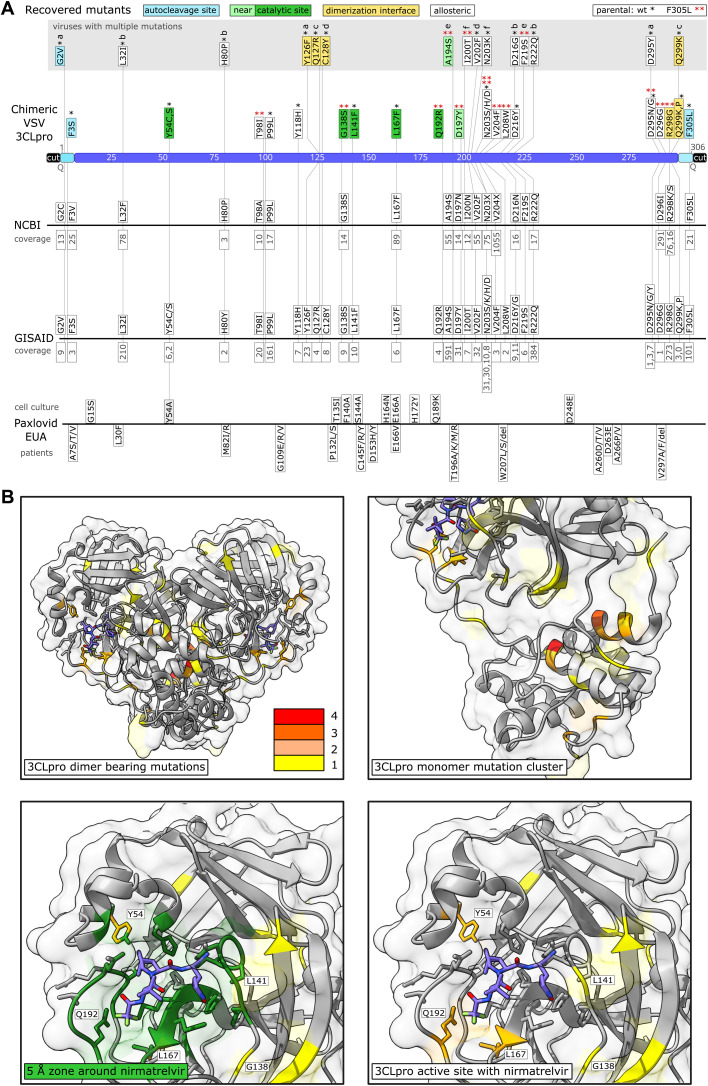
Fig. 2. Sequencing of 3CLpro escape mutants and comparison to data bases and Paxlovid EUA information.
(A) Mutants were recovered from VSV-G-3CLpro-L wild-type (*) and the F305L variant (red **). Autocleavage site mutants are colored in turquoise, catalytic site mutants in green, near catalytic site mutants in light green, dimerization interface mutants in yellow and “allosteric” mutants in white. Viruses with more than one mutation are displayed above in a gray box and named a to f. The number of mutated sequences in the databases from NCBI and GISAID are displayed below the mutations in gray. If specific mutations were not present in the database, the residue is displayed with any mutation that occurred at this position. Multiple such different amino acid changes that were not selected in our virus are displayed with X (N203X, V204X). Mutations from the Paxlovid EUA are divided into mutations found in cell culture and mutations sequenced from treated patients. The coverage of mutation entries was obtained on June second, 2022. (B) Visualizations of mutation-affected residues are shown. Residues that were mutated one time are highlighted in yellow, two times in light orange, three times dark orange, and four times in red. The 3CLpro protease dimer with bound nirmatrelvir (blue) was visualized in ChimeraX from the Protein Data Bank structure 7vh8 (32). Catalytic center mutations are within a range of 5 Å as visualized in dark green.