FIG 1.
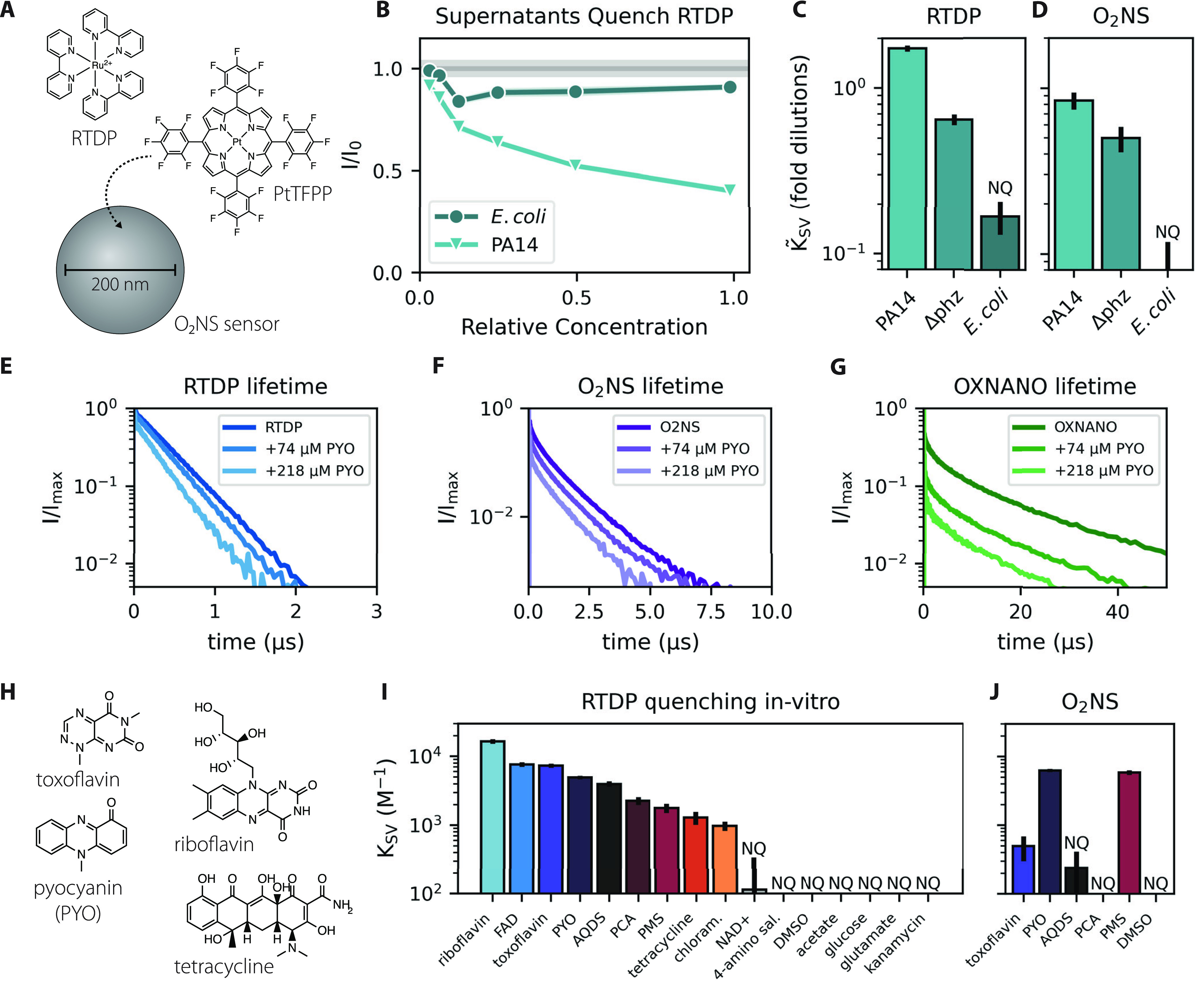
Bacteria secrete soluble quenchers of phosphorescent O2 sensors. (A) Tris(2,2′-bipyridyl)dichlororuthenium(II), or RTDP, is a soluble phosphorescent ruthenium complex that can be used as an O2 sensor (17). Platinum (II) meso-tetra(pentafluorophenyl)porphine, or PtTFPP, is the O2-sensing component of O2NS nanosensors, which encapsulate the Pt-porphyrin with a reference dye in a spherical ≈200 nm PVC matrix (13). (B) Strains were grown in a minimal medium where P. aeruginosa (PA14) reliably produces pyocyanin (26). Serially diluted PA14 supernatants quenched RTDP in a concentration-dependent manner. I/I0 denotes fluorescence normalized to no quencher, and the width of the horizontal gray bar gives the standard deviation of RTDP intensity in culture media. (C–D) Quantification shows that PA14 supernatants quench RTDP and O2NS, and that quenching is partially due to secreted phenazines. The concentration-dependence of quenching was fit to a Stern-Volmer model (19) where . Quencher concentration [Q] has units of (fold dilutions)−1 here. implies I/I0 = 0.5 when [Q] = 1, i.e., for an undiluted supernatant; larger imply stronger quenching. Deletion of phenazine biosynthesis genes (PA14 Δphz) reduced but did not eliminate quenching, and E. coli supernatants quenched both sensors far less than PA14. (E–G) Phosphorescence lifetime measurements of RTDP, O2NS and a commercial O2 sensor, OXNANO, indicated that the phenazine pyocyanin (PYO) is a strong concentration-dependent quencher of all three sensors. Panels give normalized intensities (I/Imax). (H) A subset of biological molecules tested for RTDP quenching. Panel (I) gives the results of serial dilution measurements akin to panel B. Here, KSV has M−1 units. Redox active molecules capable of spontaneous electron transfer were stronger quenchers. (J) O2NS was quenched by many of the same molecules, but encapsulation appears to protect against quenching by polar and charged species like toxoflavin and PCA. Error bars mark a 95% confidence interval. “NQ” denotes “nonquencher” where the fit to a Stern-Volmer model was poor or quenching was too weak to quantify (Fig. S1). Abbreviations: flavin adenine dinucleotide (FAD), 9,10-Anthraquinone 2,6-disulfonic acid (AQDS), phenazine 1-carboxylic acid (PCA), phenazine methosulphate (PMS), chloramphenicol (chloram.), NAD (NAD+), 4-amino salicylate (4-amino sal.), dimethyl sulfoxide (DMSO).
