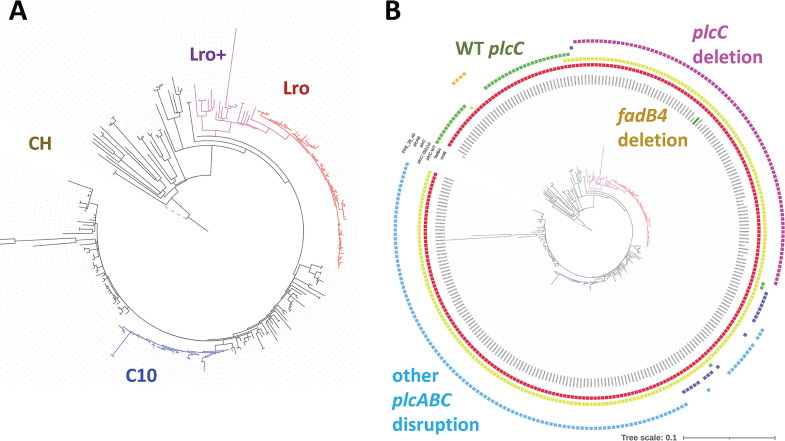
FIG 1.
Phylogenetic analysis of the genomes of M. tuberculosis lineage 3 isolated in the Midlands. (A) The Lro outbreak is a distinct cluster of 63 genomes that differ by an average of <10 SNPs (shown in red) and is part of a larger clade, which is labeled as Lro+ (purple, includes 6 genomes from isolates from Africa, whereas all other genomes are isolates from the Midlands). Genomes related to Lro were selected by the presence of the nrdB deletion, and the tree was rooted using the genome of the CH outbreak strain (dashed brown line). A second cluster has been labeled C10 (blue). (B) The Lro outbreak is characterized by 3 deletions that are unique to the Lro+ clade: nrdB, fadB4, and plcC. The presence, absence, or disruption of genes in each genome is depicted with colored boxes around the edge of the tree. Red, deletion in nrdB; green, intact plcC; yellow, disrupted fadB4. Purple, dark blue, pale blue, and orange indicate polymorphisms in or near the plcC cluster, a deletion in plcC or plcAB, an insertion, and a PPE disruption, respectively. The genome highlighted in green represents the isolate studied in Fig. 2–4.