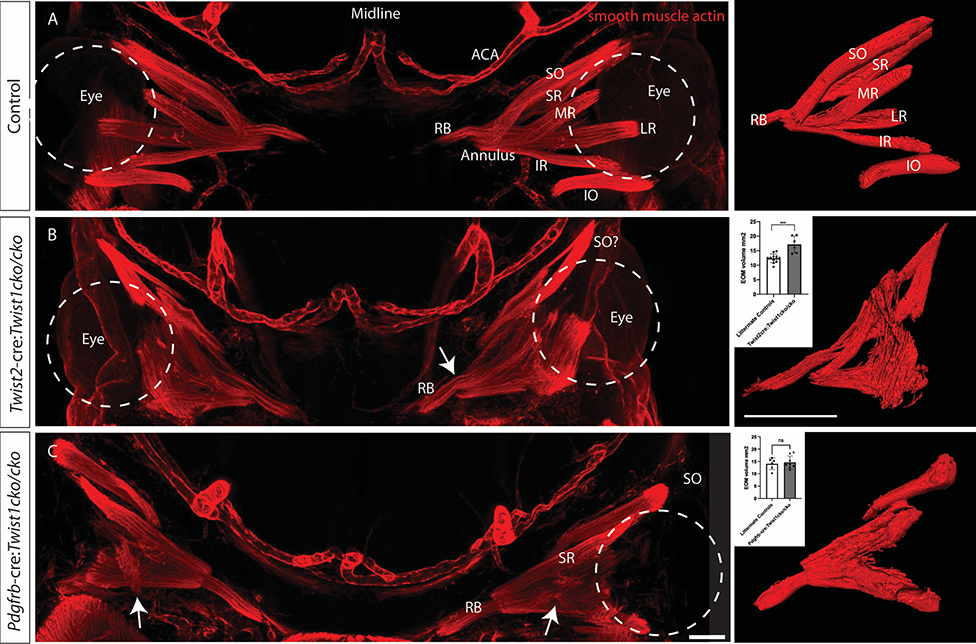
Figure 1.
Loss of Twist1 in different subsets of mesenchyme leads to varying degrees of EOM disorganization. Whole mounts from E13.5 embryos stained with anti-smooth muscle actin and imaged coronally show (A) the normal arrangement of the EOMs around the eye (dashed circle indicates left eye with red EOMs labeled) and the arteries of the anterior head. To the right of each image is a 3D reconstruction of the EOMs. (B) Loss of Twist1 in Twist2-cre expressing cells leads to profound disorganization of the EOMs. There is a muscle superiorly that appears to make a turn as if going through the trochlea, which may be the superior oblique (labeled SO?). This muscle, however, appears different from the wild-type superior oblique, in which muscle tissue extends only to the trochlea, not through it (compare to SO in panel A; in wild-type mice, a tendon (not labeled) extends from the globe to the trochlea). The RB and annulus of Zinn can be identified (arrow), but the other EOMs do not separate into distinct muscles. The total volume of EOMs is greater in the mutant animals (graph, inset). (C) Loss of Twist1 in Pdgfrb-cre expressing cells leads to mild disorganization of the EOMs. The SR, SO, and RB can be identified, but the other EOMs do not fully separate into distinct muscles, and there are muscle fibers oriented obliquely across the orbit, not originating at the apex (arrow). Consistent with the less severe phenotype, the total volume of the EOMs is not different from littermate controls (graph, inset). RB: retractor bulbi, IR: inferior rectus, MR: medial rectus, SR: superior rectus, LR: lateral rectus, SO: superior oblique, IO inferior oblique. ACA: anterior cerebral artery. Scale bar equals 200 um. n = 3–6 for each genotype. Control images are from littermate controls who do not express cre recombinase and/or have wildtype Twist1 alleles.