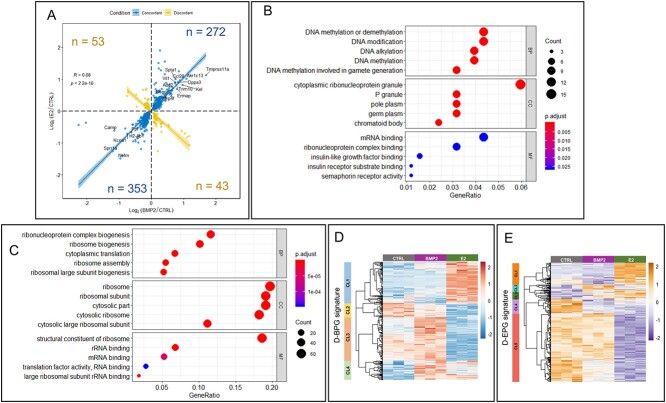
Figure 4.
Regulation of gene expression in developing ovary by both E2 and BMP2. (A) Concordant (blue dots and line) and discordant (yellow dots and line) gene expression from either BMP2 treated versus untreated control (P < 0.05) or E2-treated versus untreated control (P < 0.05). Number of genes are represented in each quadrant. Spearman correlation coefficient R = 0.68, P < 2.2e-16. (B and C) GO pathway enrichment analysis from biological process ontology (BP), cellular component ontology (CC), and molecular function ontology (MF) with both concordantly upregulated (B) and concordantly downregulated (C) DEGs in response to either BMP2 or E2 treatment versus untreated control. (D and E) Clustered heatmap illustrating expression of DEGs from either BMP2- or E2-treated ovaries that overlapped with the gene signature for differentiating bipotential pre-GC cells (D-BPGs) (D) or differentiating epithelial pre-GC cells (D-EPGs) (E). Identified major clusters (CL) are represented in the left color-coded bar. Heatmap was prepared with normalized counts from either untreated or BMP2-treated or E2-treated samples with row-centered color scheme.