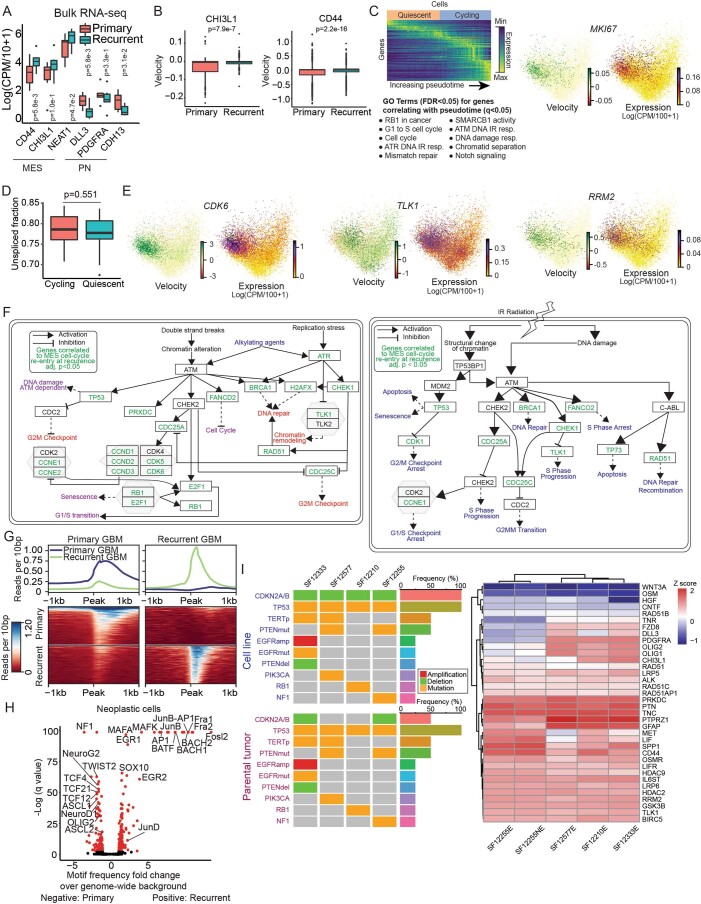
Extended Data Fig. 3. Validation, correlates, and models of the MES shift.
a) Comparison of PN, MES and monocytic-lineage cell marker genes between primary and recurrent bulk RNA-seq of patient-matched GBM longitudinal specimens, N = 30 tumors. A one-sided paired T-test was used to assess significance. Boxplots in panels A-B and D are defined as follows, lower/upper whiskers: smallest/largest observation ≥/≤ the lower/upper hinge −/+ 1.5 times the interquartile range (IQR); lower/upper hinge: 25th/75th percentile; center: 50th percentile. b) Boxplots of RNA velocities for MES hallmark genes CD44, CHI3L1 computed over PN neoplastic cells from recurrent GBM cases, N = 37,428 cells. A one-sided signed Wilcoxon rank-sum test was used to assess significance. c) (Left) A heatmap representation of the pseudotime inference shown in Fig. 3d, e is shown above. Gene ontology terms from WikiPathway.org that are over-represented (FDR < 0.05) based on genes that correlate with pseudotime, is shown below. (Right) RNA velocity and expression for MKI67 in MES neoplastic cells from recurrent GBMs, visualized in PCA space. d) The fractions of total spliced and unspliced mRNAs, compared between cycling and quiescent MES cells from recurrent cases using a two-sided Wilcoxon rank-sum test, p = 0.0551, N = 10,456 cells. e) RNA velocity and expression for CDK6, TLK1, and RRM2 in MES neoplastic cells from recurrent GBMs, visualized in PCA space, N = 10,456 cells. f) (Left) The DNA-damage response pathway adapted from Wikipathway.org. (Right) The response to IR pathway adapted from Wikipathway.org. Genes correlating with cell-cycle re-entry by MES cells at recurrence are annotated in green. g) Read-density and heatmap plots summarizing reads mapping to primary- and recurrent-specific scATAC-seq peaks respectively, from N = 10,981 cells. h) Over-represented transcription factor motifs in primary- and recurrent-specific scATAC-seq peaks, from N = 10,981 cells. i) (left) A comparison of genotypes between each of the patient-derived cell lines and the tumor specimens from which they were derived, performed via UCSF500 clinical genotyping. (right) Expression levels via RNA-seq of the patient-derived cell lines for PN and MES markers and genes targeted in in vitro assays.