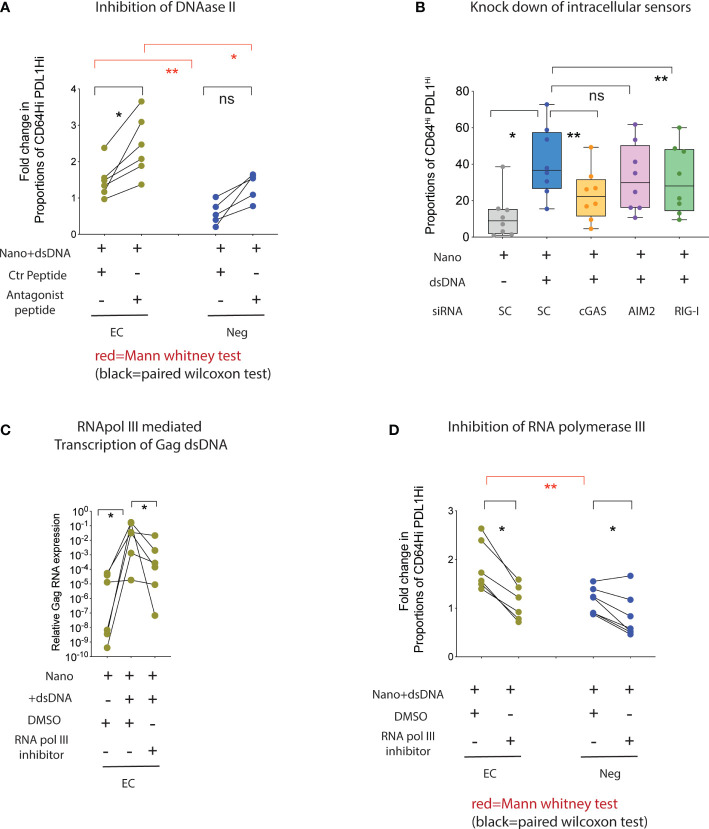
Figure 4.
Collaboration of RNA polymerase III, cGAS and RIG-I enhance the detection of intracellular HIV-1 Gag dsDNA in mDC from high responder EC. (A, D) Proportions of CD64Hi PD-L1Hi mDC from HR-EC (green) and HIV-1 negative individuals (Neg, blue) after stimulation with nanoparticles loaded with HIV-1 Gag dsDNA or control nanoparticles. Experiments were performed in the presence of either a control peptide or an antagonistic peptide against DNAase II (A) or with DMSO or an inhibitor for RNA polymerase III (D). Statistical significance was tested using two-tailed matched pairs Wilcoxon tests (black; * p<0.05) or Mann Whitney tests (red; *p<0.05; **p<0.01), respectively. (B) Fold change in proportions of CD64Hi PD-L1Hi mDC in response to stimulation with HIV-1 dsDNA after nucleofection with siRNAs specific for genes encoding the indicated intracellular nucleic acid sensors. Data were normalized to mDC nucleofected with control scramble (SC) siRNAs in the presence of HIV-1 dsDNA stimulation. Baseline normalized levels of mDC nucleofected with scramble siRNA in the absence of HIV-1 dsDNA are also included. (C) RT-qPCR analysis of HIV-1 Gag mRNA in mDC from high responder EC cultured with control nanoparticles (Nano) or nanoparticles loaded with HIV-1 gag dsDNA. Experiments were performed in the presence of DMSO or an inhibitor for RNA polymerase III. ns, not significant.