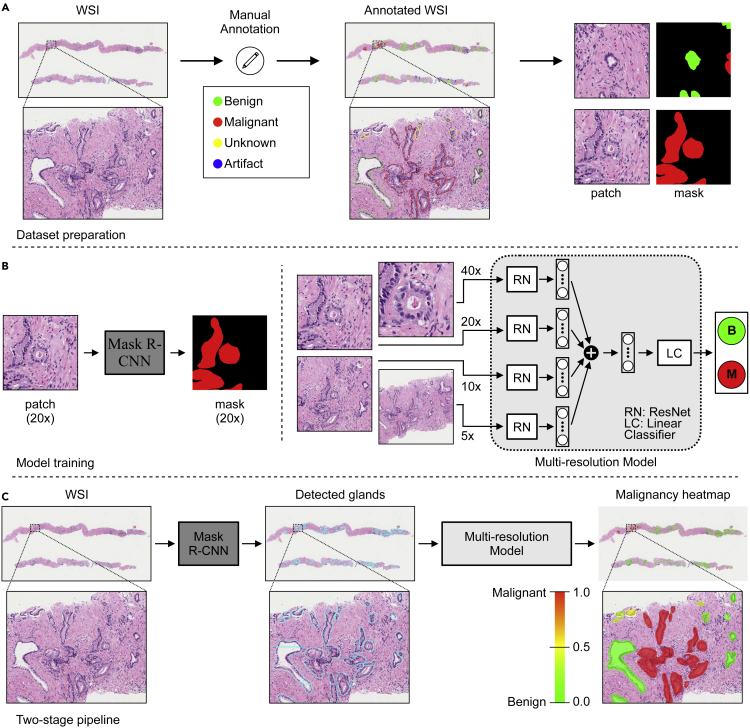
Figure 1.
Malignant gland detection pipeline in prostate core needle biopsies
(A) Glands in collected WSIs were annotated by drawing contours around each gland and assigning a malignant, benign, unknown, or artifact label. Then, patches centered around a particular gland were cropped at different resolutions for each gland while preparing datasets for training machine-learning models. See also Figure S1.
(B) A Mask R-CNN19 model was trained for gland segmentation on patch and mask pairs prepared at 20× resolution. A multi-resolution (four-resolution) deep-learning model was trained for gland classification on patches (cropped at 5×, 10×, 20×, and 40× resolutions) and label pairs.
(C) After gland segmentation and classification models were successfully trained, a two-stage pipeline was constructed. The trained Mask R-CNN model detected the glands in a WSI, and the trained multi-resolution model obtained the malignancy probability for each detected gland. Finally, a malignancy heatmap was generated to support pathologists in prostate cancer diagnosis from core needle biopsy WSIs.