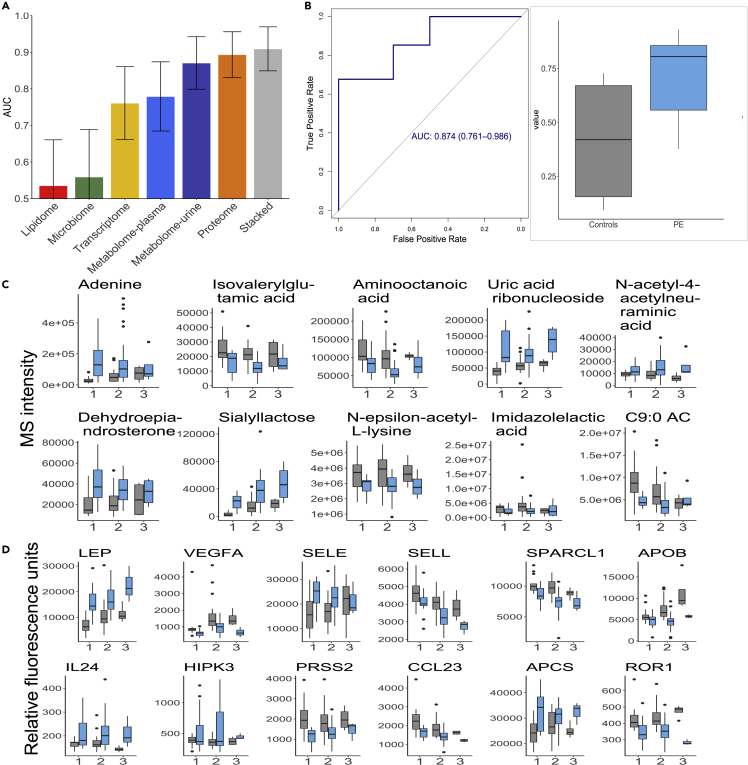
Figure 4.
An integrated multiomics machine model outperforms single omics models for preeclampsia
(A) Cross-validated performance of machine-learning models in terms of the AUC is shown on the y axis. Each model was obtained using all available samples over gestation. The integrated (stacked) model utilizing stacked regression exhibited the highest accuracy (AUC = 0.91, 95% CI [0.85, 0.97]). Both proteome and metabolome (urine) had high prediction performance (AUC = 0.89, 95% CI [0.83, 0.95] proteome; AUC = 0.87, 95% CI [0.80, 0.94] urine metabolome).
(B) Urine metabolome prediction model using ten metabolites sampled over gestation validates on the validation cohort. AUC = 0.874, 95% CI [0.76, 0.99], and prediction values (scores) obtained by EN for normotensive and preeclamptic (PE) women.
(C) Metabolites identified by EN as biomarkers of preeclampsia over gestation. y axis shows values stratified by normotensive (gray) and preeclamptic (light blue) pregnancies. p values obtained using linear mixed-effects univariate analysis show statistical significance of each metabolite (p < 0.05).
(D) Proteins identified by EN as biomarkers of preeclampsia over gestation. y axis shows a protein value stratified by normotensive (gray) and preeclamptic (light blue) pregnancies. p values obtained using linear mixed-effects univariate analysis show statistical significance of each metabolite (p < 0.05).