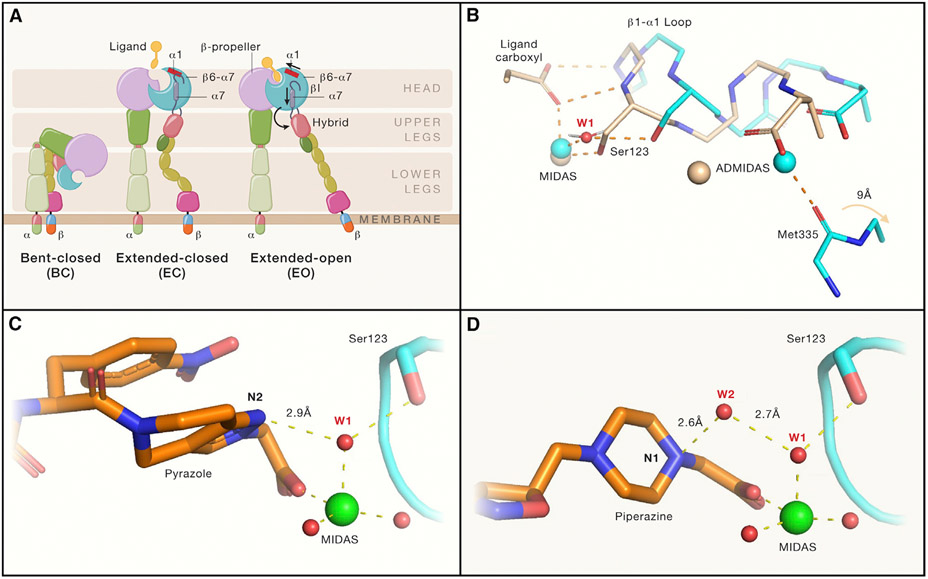
Figure 1. Integrin αIIb(β3 conformational states and mechanisms by which “closing” αIIb(β3 antagonists stabilize the closed conformation.
(A) Integrin domain organization and conformational states. Resting integrin receptors primarily adopt a bent-closed conformation, and with activation and ligand binding transition to extended-closed and extended-open conformations. Integrin headpiece opening increases affinity for biological ligands and is accomplished by ligand binding, which causes α1-helix pistoning (red bar) of the βI domain, leading to pistoning of the α7-helix (purple bar), and swing-out of the hybrid domain (curved arrow), accompanied by (B) rearrangement of loops at the ligand-binding site.
(B) Shows that the resting integrin adopts a closed conformation (cyan carbons and metals; PDB 3T3P) in which a tightly bound water molecule (water 1; W1) mediates the interaction of β3 Ser123 with the MIDAS metal ion. With the binding of the fibrinogen peptide (wheat carbons and metals; PDB 2VDO), the ligand carboxyl pulls the backbone nitrogens on the β1-α1 loop toward the MIDAS, resulting in the Ser123 now directly coordinating the MIDAS metal ion and freeing the Met335 on the β6-α7 loop to move ~9 Å (out of the frame) as part of the swing-out motion of the hybrid domain. Thus, opening of the receptor requires displacement of water 1.
(C and D) (C) The hydrogen acceptor pyrazole nitrogen of the “closing” antagonist UR-2922 stabilizes water 1 and (D) the hydrogen donor piperidine nitrogen of “closing” compound BMS4 stabilizes water 2 (W2), which in turn stabilizes water 1. Thus, both compounds prevent the displacement of water 1, which is required for reorganization of the ligand binding region and initiation of the swing-out motion. Figure adapted from Lin et al. (2022) and made with the assistance of Dr. Deena Oren of the Rockefeller University Structural Biology Resource Center.