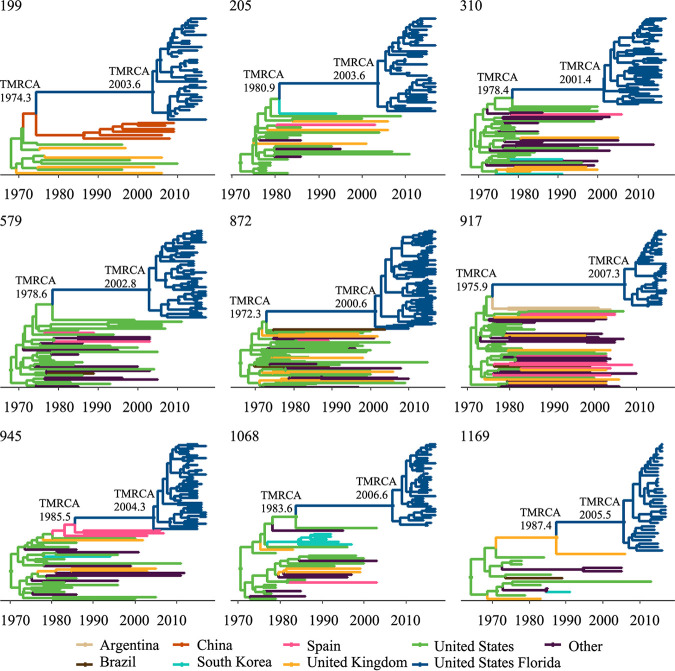
FIG 4.
Bayesian phylodynamic reconstruction of the nine largest HIV-1 subtype B clusters in Florida with reference sequences from the Los Alamos National Laboratory (LANL) database. The maximum clade credibility (MCC) time-scaled phylogenies were inferred using the relaxed molecular clock and skyline demographic priors implemented in BEAST v1.10.4, and a discrete asymmetric trait analysis. Circles represent branches supported by posterior probability >0.90. Branches are colored based on location of origin as indicated in the key (e.g., the Florida sequences in cluster #1169 share an ancestor with a sequence from the United Kingdom). Time of the most recent common ancestor (TMRCA) for Florida clusters and for the coalescent event with the most recent ancestor from LANL are indicated at the respective nodes, for 95% HPD intervals see Table S2.