FIG 1.
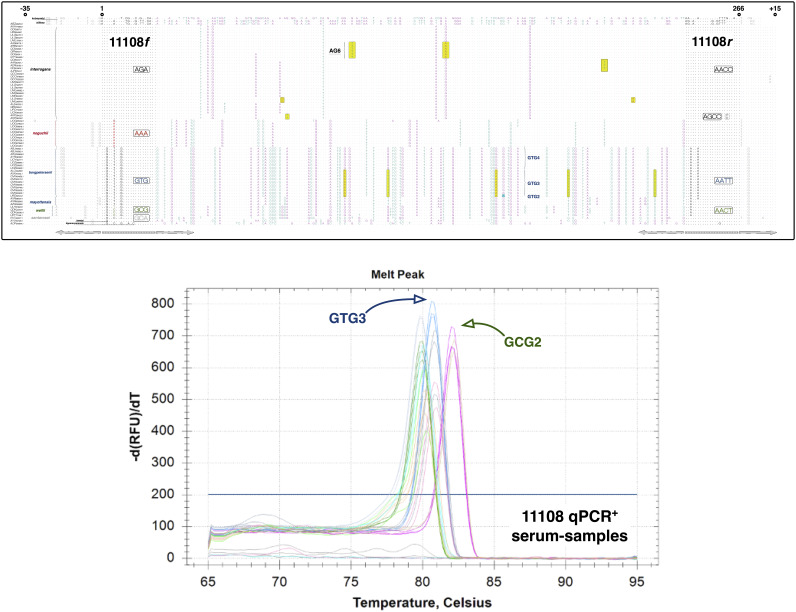
Schematic showing predicted and experimentally validated 11108-amplicon sequence-variants in patient sera. (Top panel) Global alignment of 266-bp region amplified by 11108 primer pairs with additional nucleotides at the 5′ and 3′ end, (+35 and −15, respectively). Forward (i.e., 11108f) and reverse primers and their species-associated sequence variants, e.g., 11108f_AGA for interrogans/kirschneri and noguchii strains, have been indicated. Arrows below MSA indicate alternate primer sites. Examples of allele defining SNPs are enclosed in yellow boxes. AG6 = svCopenhageni. GTG3 = borgpetersenii, unknown (UNK) serogroup (i.e., no agglutination in serotyping reactions). GCG2 = weilii UNK serogroup. AG6 is dominant human infecting strain in Iquitos (Peru), whereas GTG3 and GCG2 comprise novel noninterrogans serovars isolated from hospitalized patients in Sri Lanka—Table S3. Leptospira spp. shown: kobayashi (top, nonpathogen), biflexa (nonpathogen), interrogans, noguchii, borgpetersenii, mayottensis, weilii, santarosai, kmetyi and tipperaryanensis (bottom). (Bottom panel) CFX96-generated melt-curves (without Precision Melt Software), showing Tm’s and allele assignments from six (of 12) qPCR-positive serum-samples. Three species were reliably distinguished: interrogans (unlabeled peak), borgpetersenii GTG3, and weilii GCG2, consistent with predicted allele assignments and 11108-based amplification of cognate isolates.