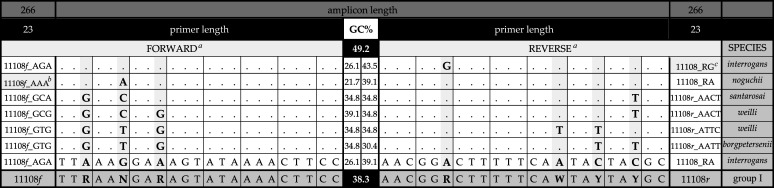
TABLE 4.
11108 qPCR assay, forward and reverse primer pairs
aThe 11108f_AGA/11108r_RA primer pair should amplify most L. interrogans, L. kirschneri, and L. noguchii alleles.
bAlternate forward primer for detection of some L. noguchii strains—especially those found in Iquitos, Peru.
cAlternate reverse primer for svManilae—unique among known L. interrogans/L. kirschneri/L. noguchii serotypes. *f_GCA/*r_AACT detect L. santarosai; *f_GCG/*r_AACT and *f_GTG/*r_ATTC (for some serotypes) detect L. weilii and *f_GTG/*r_AATT detect L. borgpetersenii and L. mayottensis (some of the latter require *f_GTG/*r_AACT). Nucleotides providing amplification specificity are indicated in bold, with alleles named via reference to these polymorphic sites. For example, AG11 = 11th sequence variant amplified by AGA forward and GTG4 = 4th amplified by GTG characteristic of L. interrogans svHardjo and L. borgpetersenii svHardjo, respectively. 11108-derived amplicons are of uniform length (266 bp), with species-dependent variations in GC%, reaching a maximum of 49.2% among L. santarosai-derived amplicons and a minimum of 38.3% shared by various alleles in L. interrogans, its sister species L. kirschneri, and genetically related L. noguchii. Primers *f_A[A/G]A (= *f_AAA and *f_AGA) and *r_[A/G]ACC (aka *r_RA and *r_RG, respectively) detect L. interrogans and its sister species, whereas *f_G[C/T][A/G] and *r_A[A/T][C/T][C/T] amplify all other disease causing group I species. None amplify group II pathogens nor nonpathogenic species. These primer combinations are not exhaustive but as far as we know, alternate forward and reverse primers represent all sequence variants found at those locations, such screening reactions employing premixed forward and reverse primer should amplify all group-I strains w/expected Tm’s.