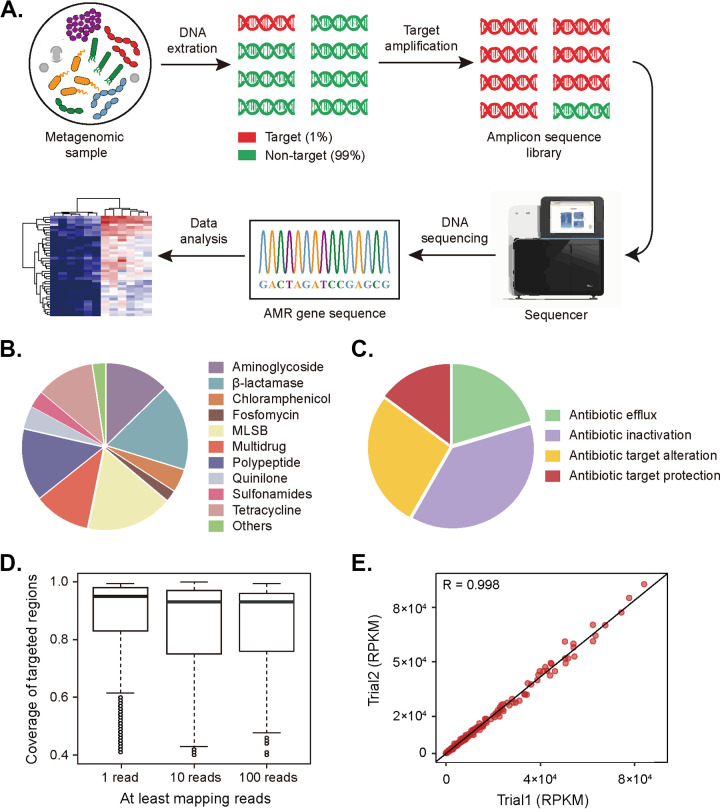
FIG 1.
Characteristic of the designed approach. (A) The whole process of amplicon sequencing. The workflow begins with DNA exaction from a metagenomic sample. Targeted sequences only account for less than 1% of the total DNA. Targeted sequences are amplified and then prepared into a sequencing library. The amplified library was sequenced, and reads were analyzed for ARGs by aligning with the sequences in the custom database for further analysis. (B) Classification of ARG determinations selected from Resfinder, CARD, and AMRFinderPlus database (n = 251). MLSB, macrolide-lincosamide-streptogramin B. (C) Classification of resistance mechanisms of selected ARG determinations. (D) Percent length of coverage of targeted regions with reads from 6 tested samples (1 versus 10 versus 100 reads). (E) Consistency of methods in different trials. Reads from samples in Trail 2 were library prep and sequenced to the same depth as reads in Trial 1. The reads were mapped to target regions, filtered for mapping quality, and then the number of reads was normalized using reads per kilobase per million (RPKM). Pearson correlation coefficients are shown in the diagram.