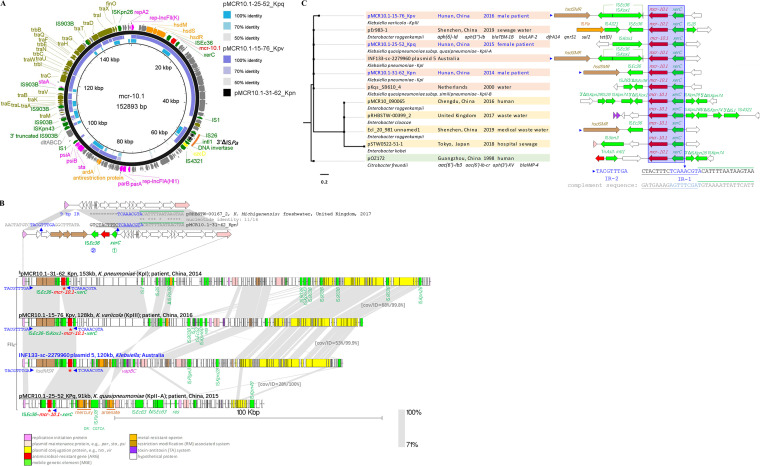
FIG 4.
Comparison of the mcr-10.1-carrying IncF plasmids. (A) Circular map showing the three mcr-10.1-carrying IncFIIK plasmids identified in this study. All plasmids were aligned to pMCR10.1-31-62_Kpn (completed by hybrid assembly; 152,983 bp). Functional genes are labeled in different colors. (B) Schematic representation showing the hsdSMR-ISEc36-mcr-10.1-xerC insertion event and comparison of the mcr-10.1-carrying IncFIIK plasmids. The 9-bp inverted repeats (IRs) are indicated in blue. The two 16-bp parts of the entire 32-bp xerC attL sequence are indicated by underlining and a green overline. Identical bases are indicated by asterisks (*). (C) Phylogenetic tree of the mcr-10.1-carrying IncF plasmids. The isolation information (source, location, and year), mcr-10.1 genetic contexts, and additional antimicrobial-resistant genes are shown. The 9-bp IRs are indicated with arrows and highlighted in blue text. The 32-bp xerC attL site is indicated by underlining and overlines. The complement sequence of attL is also shown. Δ, truncated insertion sequences.