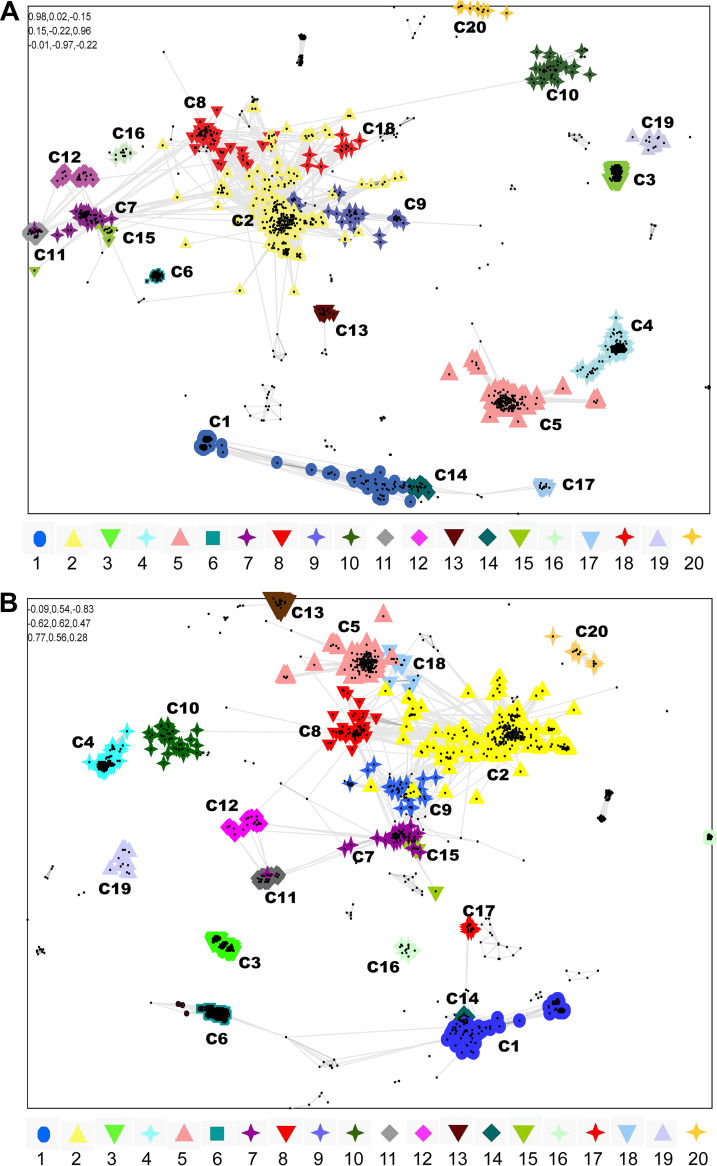
FIG 2.
Sequence similarities (CLANS) analysis-based CRESS-DNA virus capsid protein clustering. (A) A total of 1,823 amino acid sequences of Cap protein of CRESS-DNA viruses (Supplemental Data 6) were used and classified by their pairwise sequence similarity network using CLANS. The clusters were classified using the network-based method using offset values and global average with a maximum of 10,000 in CLANS Toolkit analysis. The P value of ≤1e−05 was used to show the lines connecting the sequences. (B) Pairwise sequence similarity based on CRESS-DNA virus capsid protein and +RNA viruses relationship. Representative CRESS-DNA virus capsid protein sequences and their relationship with RNA viruses using CLANS Toolkit. A total of 1,967 amino acid sequences of Cap protein of CRESS-DNA viruses and +RNA viruses were used in this analysis (Supplemental Data 8). The clusters were classified using the network-based method using offset values and global average with a maximum of 10,000 in CLANS Toolkit analysis. The P value of ≤1e−02 was used to show the lines connecting the sequences.