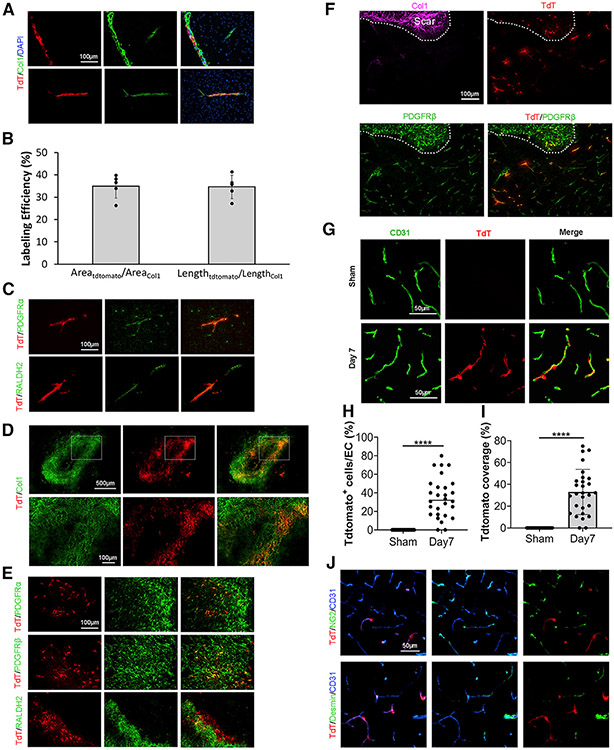
Figure 1. Lineage tracing of Col1α1+ fibroblasts.
(A) Representative images of tdTomato (TdT; red), Col1 (green), and DAPI (blue) in Col1α1-tdTomato brains under homeostatic conditions. Scale bar, 100 μm.
(B) Quantification of labeling efficiency using both vessel area and vessel length in Col1α1-tdTomato brains under homeostatic conditions. n = 5 mice.
(C) Representative images of TdT (red), PDGFRα (green), and RALDH2 (green) in Col1α1-tdTomato brains under homeostatic conditions. Scale bar, 100 μm.
(D) Representative images of TdT (red) and Col1 (green) in Col1α1-tdTomato brains at day 7 after ICH. Scale bars, 500 μm (upper panel); 100 μm (lower panel).
(E) Representative images of TdT (red), PDGFRα (green), PDGFRβ (green), and RALDH2 (green) in the peri-hematoma region of Col1α1-tdTomato mice at day 7 after ICH. Scale bar, 100 μm.
(F) Representative images of TdT (red), PDGFRβ (green), and Col1 (magenta) in the peri-hematoma region in Col1α1-tdTomato mice at day 7 after ICH. Scale bar, 100 μm.
(G) Representative images of TdT (red) and CD31 (green) in the peri-hematoma region in Col1α1-tdTomato mice in sham group and at day 7 after ICH. Scale bars, 50 μm.
(H) Quantification of the ratio of TdT-positive cells to endothelial cells in Col1α1-tdTomato brains in the sham group and at day 7 after ICH. n = 28 sections from 3 mice. ****p < 0.0001 by Student’s t test.
(I) Quantification of tdTomato coverage over CD31+ capillaries in Col1α1-tdTomato mice in sham group and at day 7 after ICH. n = 28 sections from 3 mice. ****p < 0.0001 by Student’s t test.
(J) Representative images of TdT (red), NG2 (green), Desmin (green), and CD31 (blue) in the peri-hematoma region in Col1α1-tdTomato mice at day 7 after ICH. Scale bar, 50 μm.
Data were shown as mean ± SD. ICH, intracerebral hemorrhage. See also Figures S1-S4.