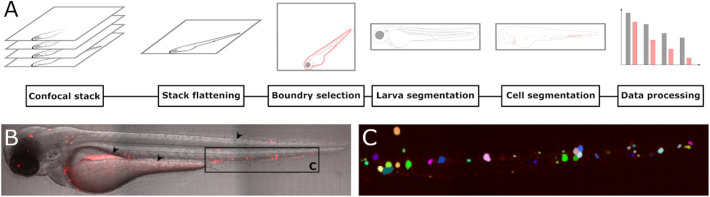
Fig. 1.
Processing of confocal images derived from zebrafish larvae intravenously injected with fluorescent cells. The workflow for segmenting cells from confocal images is illustrated in A. Acquired confocal images are first flattened to a 2D representation to enable easy visual analysis. Following flattening, the larval boundaries are selected by the user to determine the location and orientation of the larva. Using this information, the larvae are segmented and realigned to a standardised orientation. Background levels are determined by the user and all objects located within the larva are segmented using a watershed algorithm. Next, the data can be exported for further analysis. An example of the cell segmentation is given in B and C. A zebrafish larva was injected with 4 nL of a 10 106 cells·ml−1 CellTracker™ Deep Red-stained cancer cell suspension into the posterior cardinal vein at 2 dpf. A 2D representation of a confocal image acquired the day after cell injection is shown in B. Common sources of autofluorescence that need to be masked prior to segmentation are the gut, yolk sack and iridophores as indicated by black arrows. Cell segmentation of the tail region (indicated by the black rectangle in B) is displayed in C. To illustrate the segmentation, each segmented object detected is represented by a unique colour.