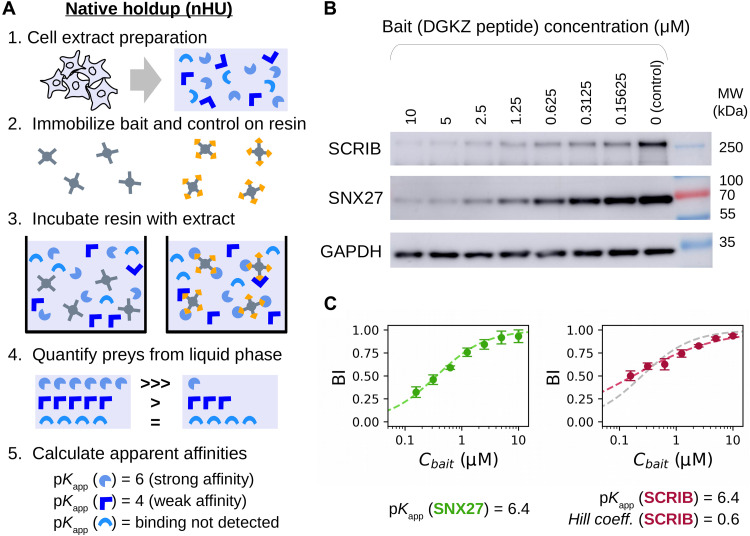
Fig. 1. Principle and simple demonstration of nHU.
(A) Schematic pipeline of nHU. Biotinylated baits and controls are immobilized on streptavidin resin at high concentration and are mixed with cellular extracts. After the binding equilibrium is reached, the liquid phase is separated by filtration or by centrifugation, and amounts of prey proteins are determined using standard protein analytical tools, such as WB or MS. The measured concentration ratio, in combination with the estimated amount of the immobilized bait concentration can be directly converted into apparent equilibrium dissociation constants (pKapp). Note that the discarded resin from step 3 can be optionally processed as a regular pull-down experiment. (B) Demonstration of nHU titration experiment using the biotinylated PBM peptide of DGKZ as bait and biotin as control. Increasing amounts of bait-saturated resins were incubated with total Jurkat extracts. Supernatant fractions were probed with specific antibodies against endogenous full-length PDZ domain–containing proteins SCRIB and SNX27. (C) Results of nHU-WB experiments presented in (B). BI values of SNX27 (green, left) and SCRIB (red, right) were first fitted with a hyperbolic binding equation (dashed line). In the case of SCRIB, a significantly better fit was obtained using the Hill equation (red dashed line) compared to a hyperbolic binding equation (gray dashed line). Determined parameters are indicated below the plots. BI values were determined on the basis of three replicates. See fig. S1 and table S1 for additional data.