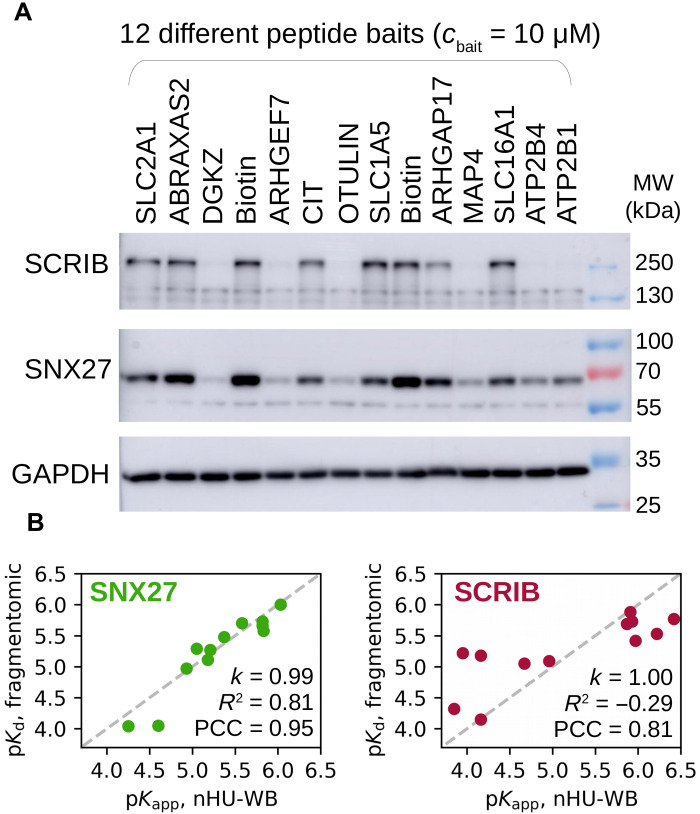
Fig. 2. Single-point nHU for rapid apparent affinity measurements.
(A) Demonstration of single-point nHU-WB using 12 different biotinylated PBM peptides (baits) or biotin (control) saturated streptavidin resin and total Jurkat extracts. Supernatant fractions were probed with specific antibodies against endogenous full-length PDZ domain–containing proteins SCRIB and SNX27. (B) Results of the nHU-WB experiment presented in (A). Correlation between in vitro fragmentomic affinities measured using PBM peptides and isolated PDZ domains (12) and apparent affinities measured with nHU between PBM peptides and full-length proteins. In the case of SCRIB (right), the site-specific fragmentomic affinities were combined assuming simple additivity. Direct proportionality was assumed between affinities (gray dashed line), and the coefficient of proportionality (k), coefficient of determination (R2) values, and Pearson correlation coefficient (PCC) values are indicated. Note the negative R2 value in the case of SCRIB, which indicates that a better fit could be obtained with a model with nonzero intercept; however, the physical basis of such model would be difficult to justify. Affinities were determined on the basis of three replicates. See fig. S1 and table S1 for additional data.