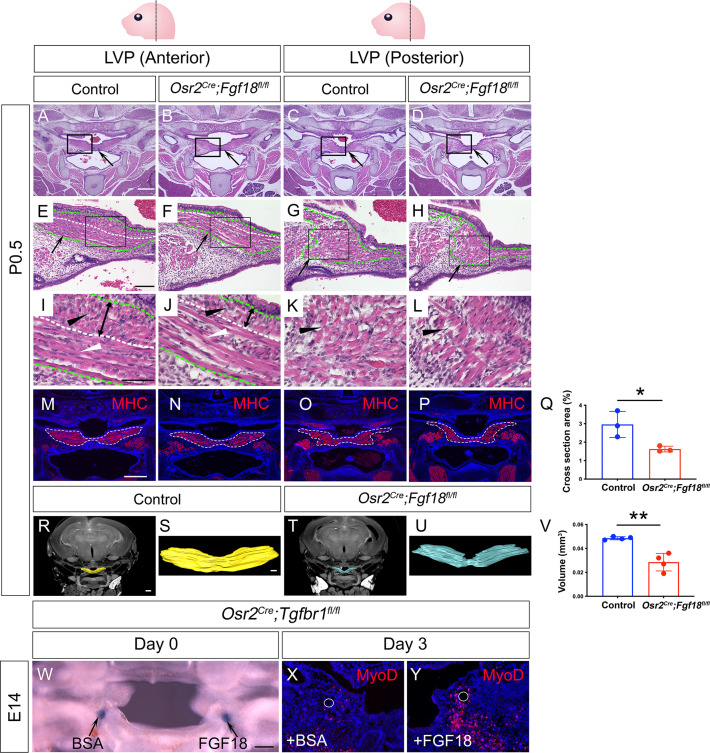
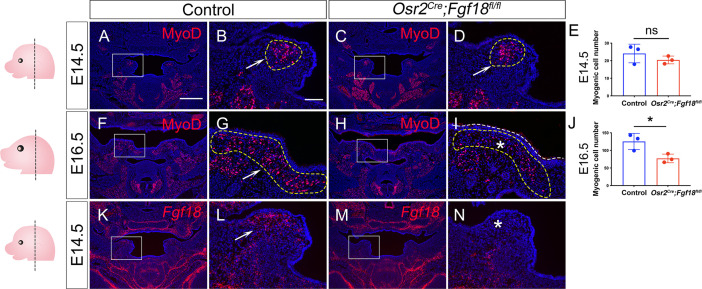
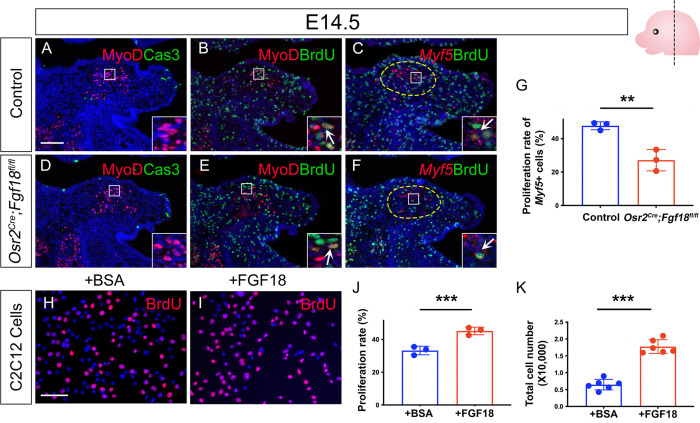
Figure 7. Myogenic cells are reduced in the LVP region of Osr2Cre;Fgf18fl/fl mice, while Fgf18 increases MyoD+ myogenic cells in Osr2Cre;Tgfbr1fl/fl soft palate slice cultures.
(A–L) H&E staining in coronal sections of LVP region at P0.5 from control and Osr2Cre;Fgf18fl/fl mice. Boxed areas in A-D and E-H are enlarged in E-H, and I-L, respectively. Black arrows point to the palatal shelf in A-D and the LVP in E-H. Green dotted line outlined the LVP in E-H. Black and white triangles point to perpendicular muscle fibers in I-L and parallel fibers in I-J. The white dotted line indicates the boundaries of perpendicular and parallel fibers in E, F, I, and J. Double-ended arrows indicate the thickness of perpendicular fibers in I and J. N=4. (M–Q) Immunofluorescence and quantification of MHC (red) staining on coronal section of the LVP region of control Osr2Cre;Fgf18fl/fl mice at P0.5. MHC+ areas in the LVP region for quantification of cross-section areas are outlined by white dashed lines in M-P. The percentage of MHC-stained area out of the whole image area is used for quantification in Q. *, p≤0.05. N=3. Statistical significance was assessed by unpaired t-test with two-tailed calculations. Data is presented as mean ± SEM. (R–V) CT scanning and quantitative analysis of the muscle volume of control and Osr2Cre;Tgfbr1fl/fl LVP at P0.5. A representative reconstructed control LVP from CT scanning is indicated in yellow (R and S) and an Osr2Cre;Tgfbr1fl/fl reconstructed LVP is indicated in teal (T and U). **, p≤0.01. N=4. Statistical significance was assessed by unpaired t-test with two-tailed calculations. Data is presented as mean ± SEM. (W) A 300 μm coronal slice of the LVP region at E14 from Osr2Cre;Fgf18fl/fl mouse for slice culture following bead implantation. Arrows point to BSA- or FGF18-treated bead. (X–Y) Immunofluorescence of MyoD (red) in the coronal section of LVP region from Osr2Cre;Fgf18fl/fl mouse cultured for 3 days with BSA bead (X) and FGF18 bead (Y). White circles indicate the location of the BSA beads in X and FGF18 beads in Y. N=3. Fgf18fl/fl or Fgf18fl/+ littermates were used as controls for Osr2Cre;Fgf18fl/fl mice. Scale bars in A, E, I, M, R, S, and W indicate 500 μm in in A-D, 100 μm in E-H, 50 μm in I-L, 400 μm in M-P, 400 μm in R and T, 100 μm in S and U, and 100 μm in W-Y, respectively.