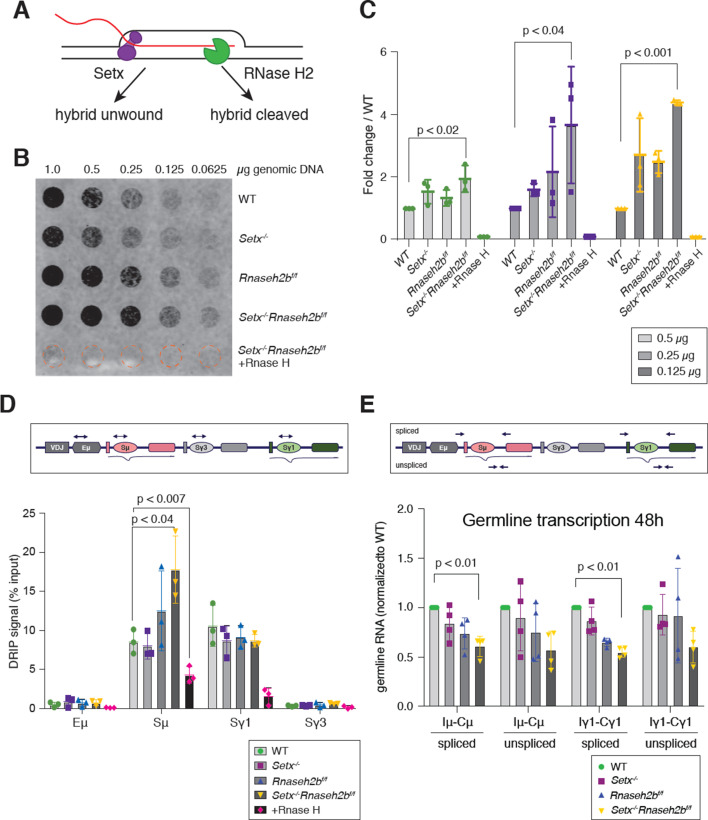
Figure 1. RNA-DNA hybrids are increased in Setx-/-Rnaseh2bf/f B cells during class switch recombination (CSR) to IgG1.
(A) Senataxin (SETX) and RNase H2 contribute to RNA:DNA hybrid removal; SETX helicase activity unwinds the nucleotide strands retaining both the RNA and DNA components while RNase H2 cleaves RNA, retaining only the DNA strand. (B) Dot blot analysis of R loop formation: twofold serial dilutions of genomic DNA starting at 1 µg were arrayed on a nitrocellulose membrane and probed using the S9.6 antibody; RNase H treatment of the Setx-/-Rnaseh2bf/f sample was used as the negative control. (C) Quantitation of 0.5 µg, 0.25 µg, and 0.125 µg dot blot from (B) using ImageJ; values were normalized to WT, set as 1. Figures are expressed as fold change relative to WT. Error bars are the standard deviation between different experiments; *p<0.05 comparing different genotypes using multiple t-test (n = 3 mice/genotype). (D) Diagram of the IgH locus showing the location of PCR products used for chromatin immunoprecipitation (ChIP) and DNA:RNA hybrid immunoprecipitation (DRIP) assays. DRIP assay was performed with S9.6 antibody using primary B cells after 72 hr of stimulation to IgG1 with LPS/IL-4/α-RP105; RNase H treatment of Setx-/-Rnaseh2bf/f sample was used as the negative control. Relative enrichment was calculated as ChIP/input, and the results were replicated in three independent experiments. Error bars show standard deviation; statistical analysis was performed using one-way ANOVA (n = 3 mice/genotype). (E) Diagram of the IgH locus showing the location of PCR products for germline transcription under IgG1 stimulation with LPS/IL-4/α-RP105. Real-time RT-PCR analysis for germline transcripts (Ix-Cx) at donor and acceptor switch regions in WT, Setx-/-, Rnaseh2bf/f, and Setx-/- Rnaseh2bf/f splenic B lymphocytes cultured for 48 hr with LPS/IL-4/α-RP105 stimulation. Expression is normalized to CD79b and is presented as relative to expression in WT cells, set as 1. Error bars show standard deviation; statistical analysis was performed using multiple t-test (n = 4 mice/genotype).