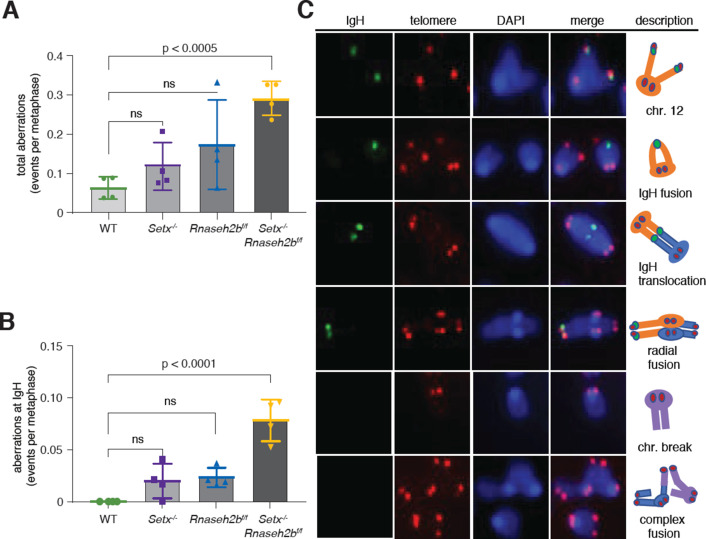
Figure 3. Increased IgH damage is observed in Setx-/-Rnaseh2bf/f B cells.
(A) Frequency of spontaneous DNA damage in WT, Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f cells. (B) Frequency of spontaneous DNA damage at IgH. (C) Representative images of the types of rearrangements produced. IgH-specific probe visualized in green, Telomere-specific probe visualized in red, DAPI is in blue. All cells were harvested 72 hr post-stimulation to IgG1 with LPS/IL-4/α-RP105. Error bars show standard deviation; statistical significance versus WT was determined by one-way ANOVA (n = 4 independent mice).