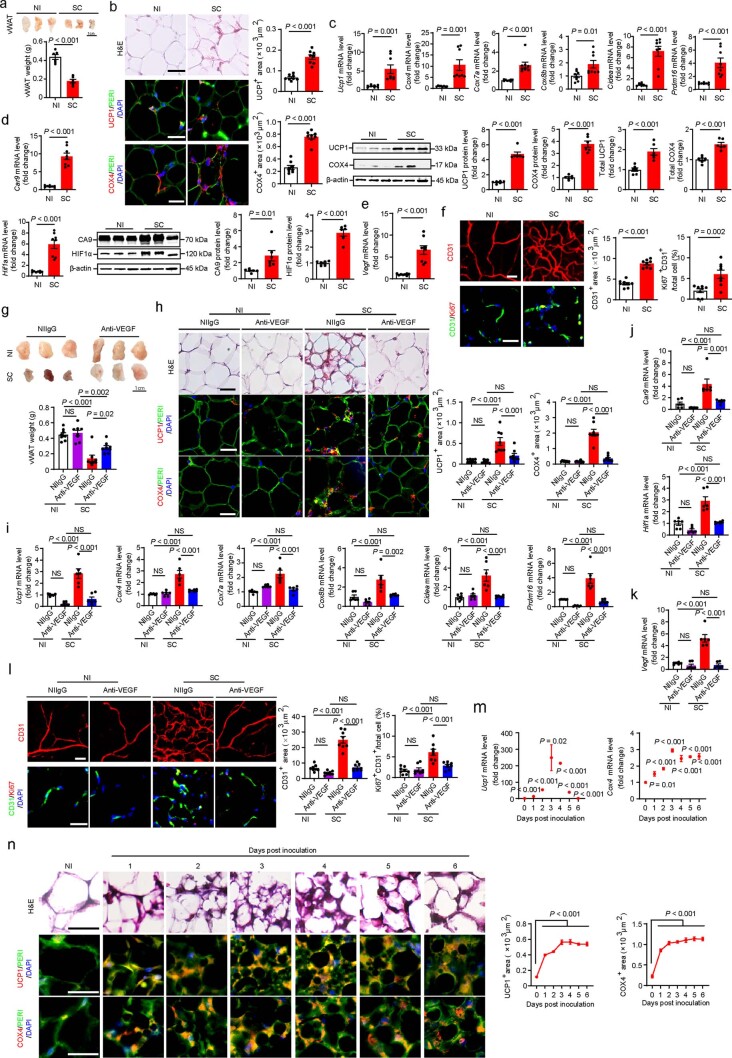
Extended Data Fig. 5. VEGF-dependent mechanisms of SARS-CoV-2-induced adipose browning.
(a) Representative pictures and weight of vWAT and vWAT weight of NI- and day-6-post SC-infected mice (n = 6 samples per group). (b) Histological and immunohistochemical analyses of vWAT of NI- and day-6-post SC-infected mice by staining with H&E, Perilipin (PERI), UCP1 and COX4. Quantification of UCP1 and COX4 positive signals in vWAT (n = 8 random fields per group). (c, d, e) Quantification of Ucp1, Cox4, Cox7a, Cox8b, Cidea, Prdm16, Car9, Hif1a and Vegf mRNA level (n = 8 samples per group); UCP1, COX4, HIF1α and CA9 protein level in vWAT of NI- and day-6-post SC-infected (n = 6 samples per group). (f) CD31 and Ki67 staining and quantification of microvessels in vWAT of NI- or day-6-post SC-infected mice (n = 8 random fields per group). (g) Representative pictures of vWAT and vWAT weight of NIIgG- and anti-VEGF-treated NI- or day-6-post SC-infected mice (n = 8 samples per group). (h) Histological and immunohistochemical analyses of vWAT of NIIgG- and anti-VEGF-treated NI- or day-6-post SC-infected mice by staining with H&E, Perilipin (PERI), UCP1 and COX4. Quantifications of UCP1 and COX4 positive signals in vWAT (n = 8 random fields per group). (i, j, k) Quantification of Ucp1, Cox4, Cox7a, Cox8b, Cidea, Prdm16, Car9, Hif1a and Vegf mRNA levels in vWAT of NIIgG- and anti-VEGF-treated NI- or day-6-post SC-infected mice (n = 6 samples per group). (l) CD31 and Ki67 double staining and quantification of microvessels in vWAT of NIIgG- and anti-VEGF-treated NI- or day-6-post SC-infected mice (n = 8 random fields per group). (m) Quantification of Ucp1 and Cox4 mRNA levels in vWAT of SC-infected mice at various time points (n = 4 samples per group). (n) Histological and immunohistochemical analyses of vWAT in SC-infected mice at various time points by staining with H&E, perilipin (PERI), UCP1 and COX4. UCP1+ and COX4+ signals were quantified (n = 8 random fields per group). Data are presented as means ± s.e.m. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey multiple-comparison test and two-sided unpaired t-tests. NS = not significant. NI = non-infected. SC = SARS-CoV-2. Scale bar = 50 μm.