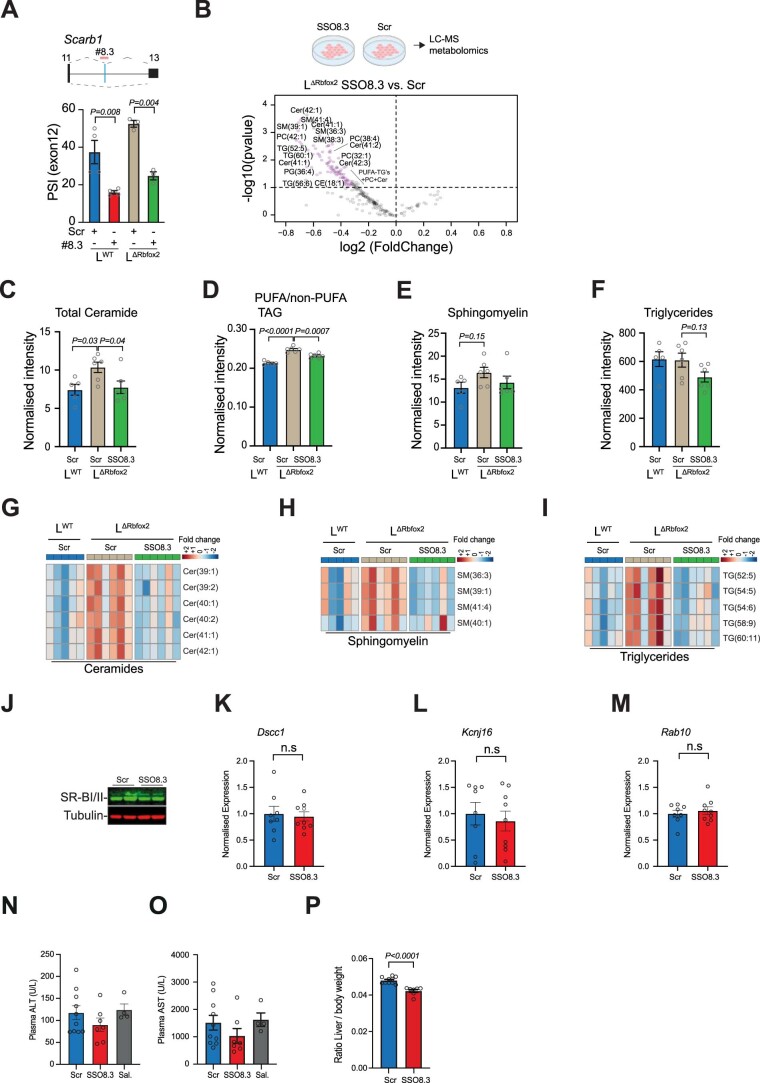
Extended Data Fig. 9. Role of Scarb1 splicing variants in lipid metabolism.
A.- Splice-switching oligonucleotide (SSO8.3) promotes skipping of Scarb1 exon 12 in primary hepatocytes as determined by semi-quantitative PCR analysis (n = 3-4). B.- Volcano-plot showing LC-MS lipidomic analysis of LΔRbfox2 hepatocytes treated with 100 nM SSO8.3 or Scr for 16 h (n = 5-6). C.- Metabolomics analysis showing levels of total ceramide, D.- PUFA/non-PUFA TG ratio, E.- total sphingomyelin, and F.- total triglycerides (n = 5-6). G-I.- Heatmap showing LC-MS metabolomic analysis of LWT and LΔRbfox2 hepatocytes treated with SSO8.3 or Scr. Intensities were normalised to internal standard and total cell counts. J.- Western-blot showing SR-BI/II levels in the liver after SSO8.3 treatment (n = 2). K.- RT-qPCR expression analysis in liver of mice treated with SSO8.3 or Scr of potential off-targets genes such as Dscc1, (L.-) Kcnj16 and (M.-) Rab10. N.- Effect of SSO8.3 injection on circulating ALT and O.- AST levels. P.- Liver/body weight ratio upon SSO8.3 injection (n = 7-10). Results are represented as mean ± s.e.m. Statistical significance was determined by one-way ANOVA or two-sided unpaired t-test of biologically independent samples.