Figure 5.
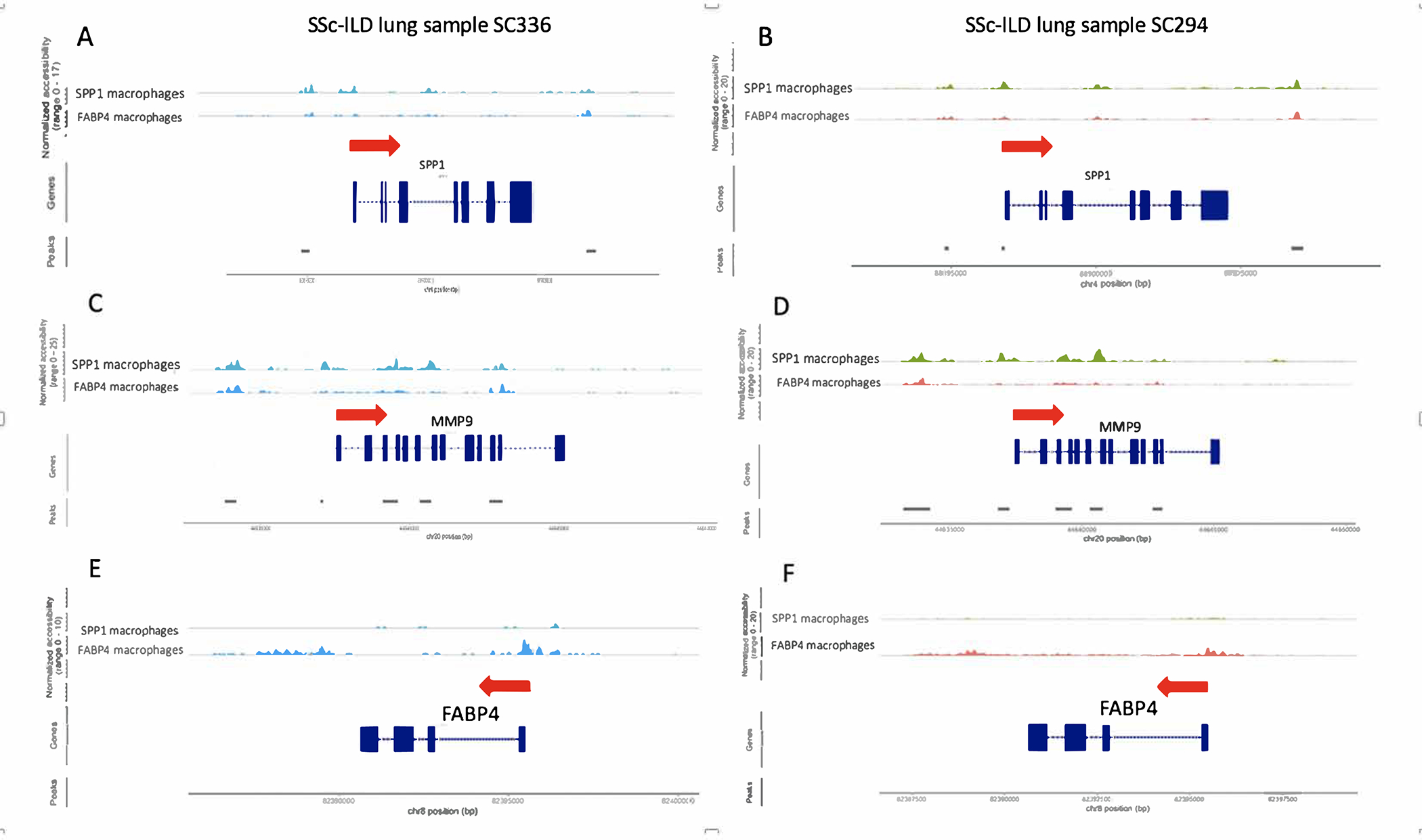
ScATAC-sequencing analysis of 2 SSc-ILD lung samples by Signac showing chromatin pattern changes for SPP1 macrophages compared to FABP4 macrophages. The red arrow indicates the direction of transcription. Exons are shown in blocks and introns flank exons. (A, B) SPP1 gene showed more accessible chromatin for the SSc-ILD SPP1 Mφ compared to FABP4 Mφ in the region proximal to the transcriptional start site for a SSc-ILD lung, as well as regions further 5’ of the promoter and in intron 4. (C, D) MMP9 gene also showed more accessible chromatin in SPP1 Mφ, but in this case increased accessibility was not seen around the promoter but rather in regions around exon 6, exons 9–12 and introns. (E, F) FABP4 gene showed strikingly more accessible chromatin in a region proximal to the transcriptional start site and in a broad second region 3’ from the gene in FABP4 Mφ compared to SPP1 Mφ.
