Figure 3: Overview of technologies for barcoding single cells.
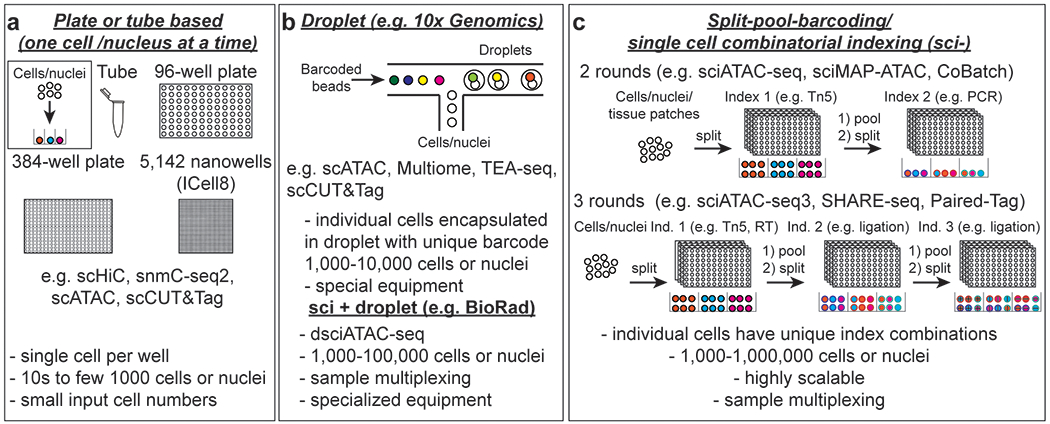
a In plate, tube, microfluidicsor nanowell chip-based assays, single cells are dispensed into individual wells or tubes or captured in reaction chambers where library preparation and molecular barcoding are carried out. These approaches usually have low throughput but can yield high coverage libraries. Plate and tube-based assays are well suited for rare cell types or assays that require high coverage such as DNA methylation and single-cell Hi-C. Throughput for plate-based assays can be increased using liquid handling robotics. IFC: Integrated fluidic circuit b Droplet-based assays allow ten thousand cells or nuclei to be profiled in parallel (left). An initial sample indexing step allows sample multiplexing prior to loading. If samples are indexed at the fragment level, channels can be superloaded to enable profiling of large numbers of cells for one sample or multiplexing of many samples. If a droplet contains more than one nucleus, sequencing reads can be assigned to individual samples or sublibraries with the initial sample index sequence.(right). Both sample and cell barcodes are used to assign reads to specific cells or nuclei. c Single-cell combinatorial indexing (sci-) or split-pool barcoding assays provide very high scalability and enables sample multiplexing by introducing a sample barcode in the first indexing round. After each indexing step nuclei are pooled and distributed to another set of plates for a total of 2 or more rounds. The cell barcode is composed of the combination of indexes from each round. With automation this approach delivers high data quality and reproducibility. RT: reverse transcription.
