Figure 4.
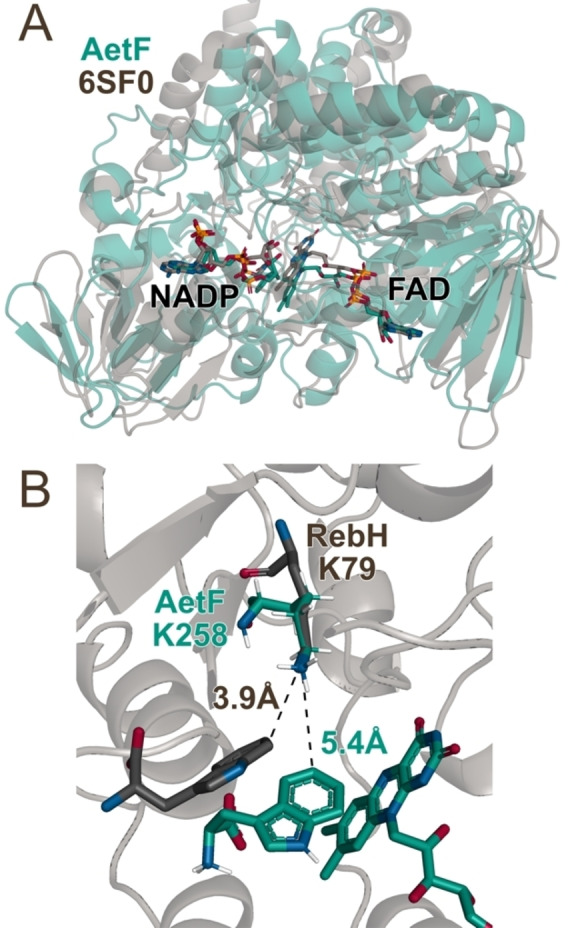
Overlays of A) a AetF⋅FAD⋅NADP model constructed using AlphaFold with cofactors docked in using Rosetta Ligand (green) with the crystal structure of a predicted ancestral FPMO (PDB ID 6SF0, grey) [32] and B) the AetF⋅FAD⋅Trp model active site (green residues) with K79 and bound Trp substrate from the FDH RebH (PDB ID 2OA1, grey residues). [34]
