Figure 2. RSL3 and ML162 specifically bind to Sec46 and Cys66 of GPX4.
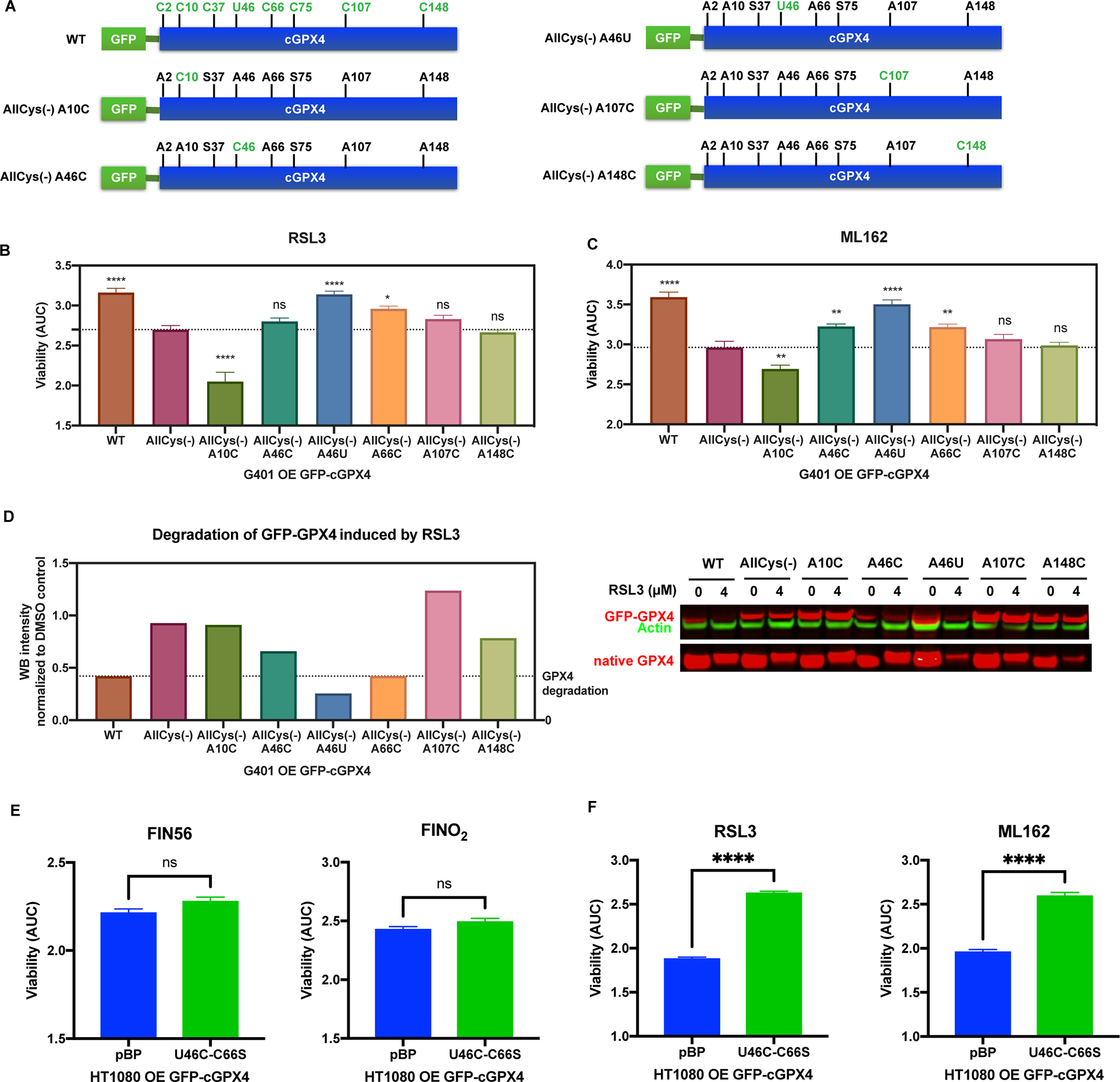
A, Construction of GFP-tagged AllCys(−) A10C, AllCys(−) A46C, AllCys(−) A46U, AllCys(−) A107C, and AllCys(−) A148C GPX4. B and C, G401 cells overexpressing exogenous WT or variants of GFP-GPX4 were tested for RSL3 and ML162 sensitivity (n=3). One-way ANOVA followed by Dunnett’s multiple comparisons test (compared to AllCys(−)) was performed: pns>0.05, p* < 0.05, p** < 0.01, and p**** < 0.0001. D, The native GPX4 along with WT and variants of GFP-GPX4 expressed in G401 was tested for vulnerability to the degradation induced by RSL3. E and F, HT-1080 overexpressing exogenous U46C-C66S GFP-GPX4 and a control line were tested for FIN56, FINO2, RSL3, and ML162 sensitivity (n=3). Unpaired t test was performed: pns>0.05 and p**** < 0.0001.
For B, C, E, and F, data are represented as mean ± s.d. See also Figure S2.
