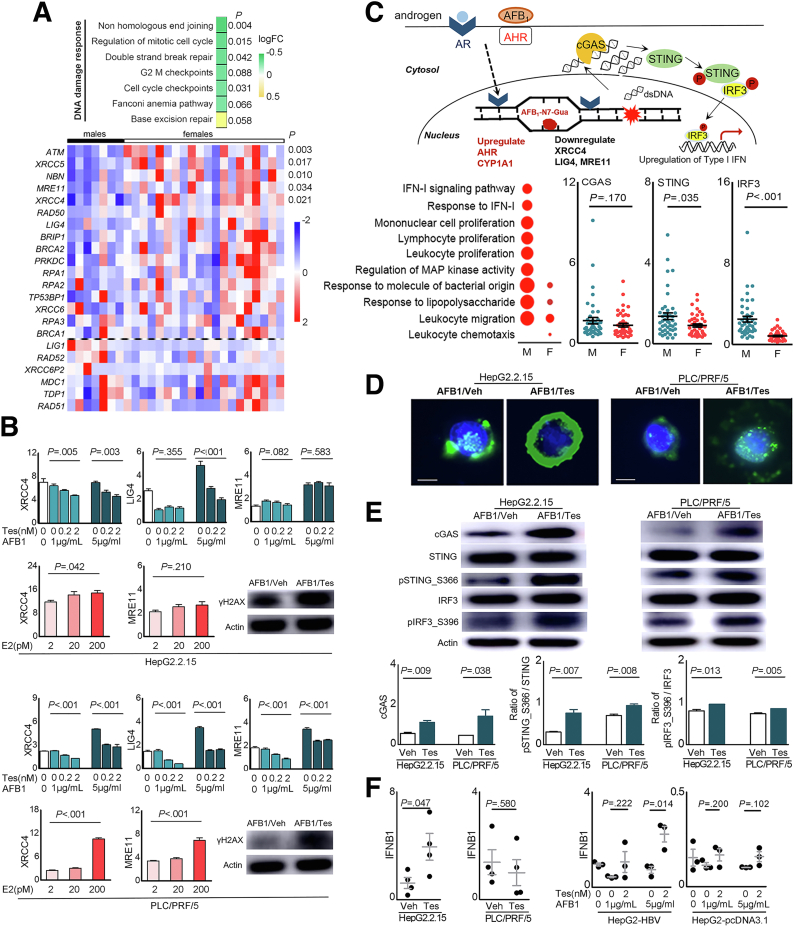
Figure 9.
Sex hormones on expression of the genes related to DNA damage responses and cGAS-STING activation. (A) The NHEJ pathway genes differentially expressed in male and female QD-HCCs that were confirmed with HBV integration. (B) Bar graphs show transcriptional levels of some NHEJ genes in HepG2.2.15 cells and PLC/PRF/5 cells that were treated with AFB1 in the presence of different concentrations of Tes, or 17β-estradiol (E2), determined by real-time quantitative polymerase chain reaction with glyceraldehyde-3-phosphate dehydrogenase as control. Images show the γH2AX expression in total cellular proteins analyzed by immunoblot of 3 independent experiments. (C) The schematic effects of AFB1 plus androgen on HCC cells. Graphs show the results of QD-HCC samples. The augmented genes related to innate immune responses analyzed by Gene Ontology are shown in the bubble graph. Bubble sizes indicate variated numbers of related genes in the specified pathway, the biggest includes 25 genes and the smallest includes 4 genes. Scatter plots show the specified genes expressed in male and female QD-HCCs. (D and E) HepG2.2.15 cells and PLC/PRF/5 cells were treated for 72 hours with AFB1 (CC50 concentration) plus 2 nmol/L testosterone (AFB1/Tes) or with the same concentration of AFB1 alone (AFB1/Veh). (D) Representative images (×400) show the cytosolic double-strand DNA analyzed by fluorescent microscopy. Scale bars: 10 μm. (E) Analysis of cGAS-STING pathway activation in differently treated HepG2.2.15 cells and PLC/PRF/5 cells by immunoblot. Images show 1 representative of 3 independent experiments, with actin as the loading control. Bar graphs show the band density of the specific genes. (F) The transcriptional levels of interferon β (IFNB1) in the cells of HepG2.2.15, PLC/PRF/5, and HepG2 transfected with 1.3 × HBV plasmid (HepG2-HBV) or empty-vector (HepG2-pcDNA3.1), described in PMID: 36058909. All cells were treated as described in panel E, with glyceraldehyde-3-phosphate dehydrogenase as control. Data represent means ± SEM. Each dot represents 1 independent experiment. P values were calculated by an unpaired Student t test. logFC, log2 fold change.