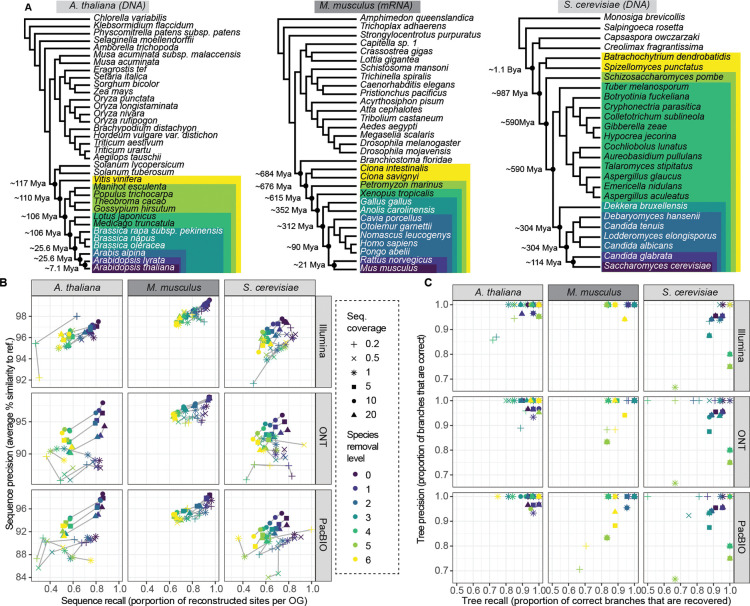
Figure 2. Benchmark of Read2Tree using three different datasets, six different coverage levels, and three sequencing technologies.
(A) Phylogenetic trees of reference datasets. In dark purple (bottom) species used for mapping. Colours represent species removal to assess dependency on closest neighbours in reference datasets. Timepoints were obtained from timetree.org. (B) Read2Tree sequences are more similar (percentage identity) and more complete with increasing coverage and decreasing distance to a more closely related species. Best sequence identity is obtained for Illumina data. Colours convey the increasing evolutionary distance to the closest reference species. (C) Precision and recall of trees reconstructed using Read2Tree after collapsing branches below 90% support.