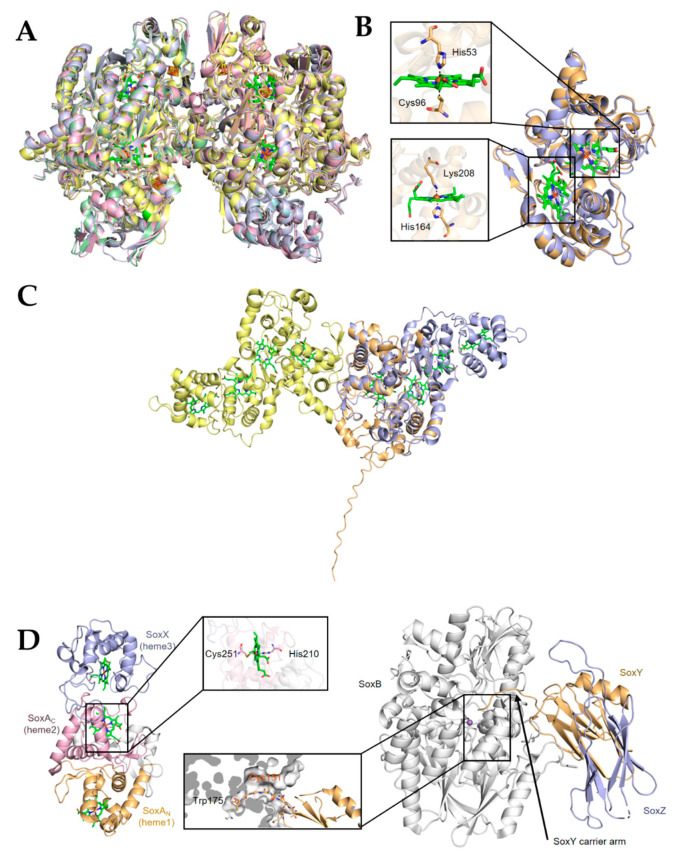
Figure 3.
Structure of representative enzymes involved in transformation of sulfur compounds in bacteria. (A) Overall structure of the Dsr A2B2 heterotetramer of Desulfovibrio vulgaris (PDB ID: 2V4J), green; Archaeoglobus fulgidus (PDB ID: 3MM5), yellow; Desulfovibrio gigas (PDB ID: 3OR1), pink; and Desulfoicrobrium norvegicum (PDB ID: 2XSJ), blue. Heme ligands are shown in ball-stick model. (B) Structure of TsdA of Marichromatium purpuratum (PDB ID: 5LO9; violet) and of Allochromatium vinosum (PDB ID: 4WQ7; orange). Heme ligands are shown in ball-stick model. Expanded regions show the ligands to hemes in active centers. (C) TsdA of Campylobacter jejuni (orange, prepared from AlaphaFold database [90,91]) is aligned and superimposed onto TsdAB of M. purpuratum (PDB ID: 5LO9), which are in violet and yellow, respectively. Heme ligands are shown in ball-stick model. Expanded region shows the ligands to heme 2. (D) Structure of the SoxXA (PDB ID: 2C1D) (left) and SoxYZ-B (PDB ID: 4UWQ) (right) complexes. Heme ligands are shown in ball-stick model. Expanded region shows the active site positioning at the substrate channel of SoxB.