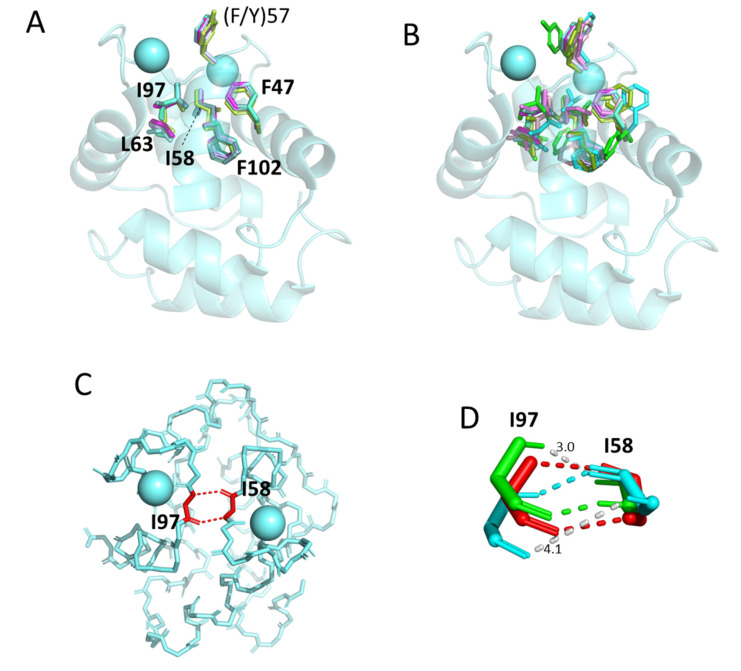
Figure 6.
The conserved core residues in the proximity of the Ca2+ binding sites show a well-conserved orientation among parvalbumin structures determined by X-ray crystallography but less so among structures determined by NMR. (A) Superimposition of X-ray crystallography structures of α-parvalbumin (magenta, spotless smooth-hound shark SPV-I [5ZGM]), oncomodulin (limon, rat oncomodulin [1OMD]), and other parvalbumins (soft purple, chicken ATH [3FS7]; green cyan, pike pvalb3 [1PVB]; aquamarine, common carp pvalb4_[Chr.A3] [4CPV]; light teal, spotless smooth-hound shark SPV-II [5ZH6]). (B) As (A), but with additional superimposition of structures determined by solution NMR of α-parvalbumin (light pink, human α-parvalbumin [1RK9]; pink, pike pvalb7 [2PAS]), oncomodulin (green, human oncomodulin [1TTX]; splitpea green, chicken oncomodulin [CPV3] [2KYF]), and other parvalbumin (cyan, Atlantic cod pvalb2 [2MBX]). (C) The beta-strand type connection between conserved residues I58 and I97 is well-conserved between all investigated structures determined by X-ray crystallography and most structures determined by solution NMR. The structure shown here, only showing the main chains in sticks format, is of common carp pvalb4_(Chr.A3) (4CPV). (D) In the solution NMR structures for human oncomodulin (1TTX) (green) and Atlantic cod pvalb2 (2MBX) (cyan), the I58 and I97 orientations are somewhat distorted compared to other investigated structures (here exemplified by common carp pvalb4_[Chr.A3] [4CPV] in red; polar contacts are indicated with dashed lines in a color matching the relevant structure depiction) and software only recognizes one and not two polar bonds between the main chains. The gray dashed lines, measured in Å, indicate distances between relevant 1TTX and 2MBX atoms where in other parvalbumin structures polar contacts are recognized.