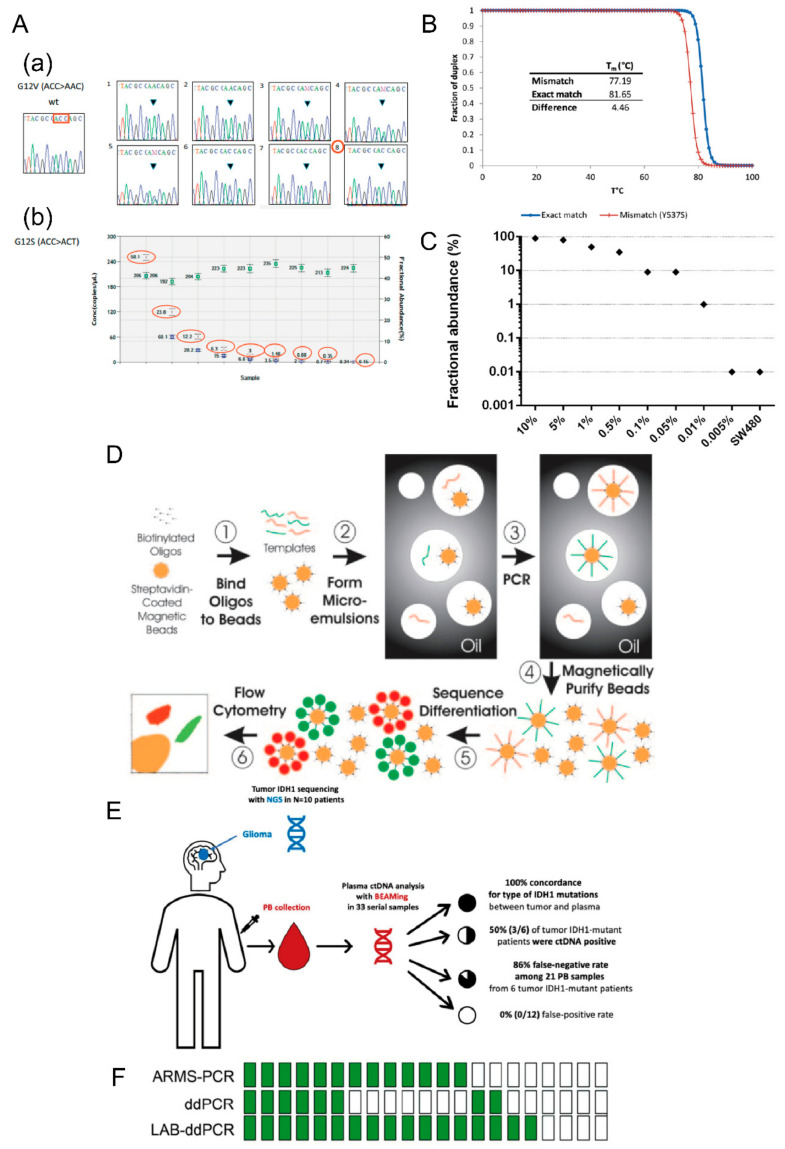
Figure 2.
Detection technology of ctDNA mutation. (A) Sensitivity of COLD-PCR (a) and ddPCR (b) for G12V-mutated DNA of metastatic colorectal cancer patients [59]. Aa, fast COLD-PCR sequence profiles of G12 V mutated DNA serially diluted with wild-type DNA (1 = 12.5%, 2 = 6.25%, 3 = 3.12%, 4 = 1.56%, 5 = 0.78%, 6 = 0.39%, 7 = 0.2%, 8 = 0.1% of mutated DNA). The antisense sequence is shown. Ab, sensitivity of the ddPCR G12S assay in discriminating different proportions of mutated alleles on serial dilutions starting from 50% up to 0.1% of the mutated allele. The respective percentages of fractional abundance obtained for each point are circled in red. (B) Melting curves and melting temperatures (Tm) of the wild-type ESR1 and ESR1 Y537S mutation [61]. (C) Sensitivity of enhanced-ice-COLD-PCR assay for ESR1 [60]. (D) Schedule of BEAMing [63]. (E) Comparison of BEAMing and NGS in detecting IDH mutation in glioma patients [65]. (F) The sensitivity of LAB-ddPCR for the detection of ctDNA T790M mutation was further improved compared to ddPCR and ARSM-PCR [68].