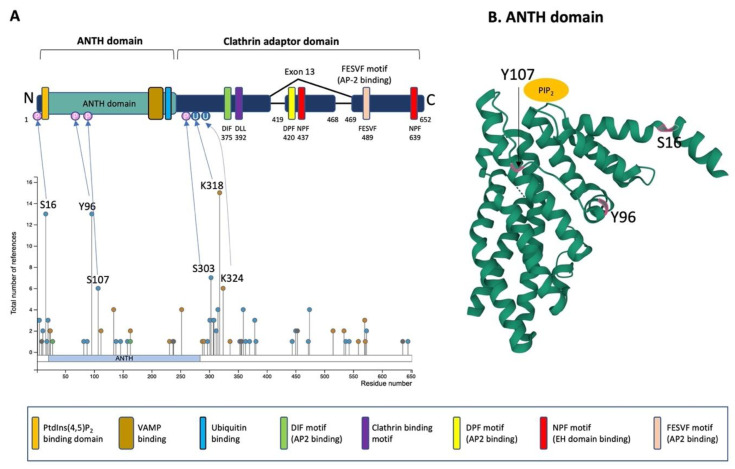
Figure 3.
Schematic illustration of PICALM structure and post-translational modifications. (A) Schematic structure of PICALM. PICALM is constituted of two distinct domains: the ANTH (AP180 N-terminal homology) domain and clathrin adaptor domain. ANTH domain contains a PtdIns(4,5)P2 binding domain and vesicle-associated membrane protein (VAMP) binding domain. Clathrin adaptor domain contains several motifs that are critical to interact with endocytic protein, including DIF, DLL Clathrin binding, DPF, NPF and FESVF motifs. DIF motif binds to AP2. DLL motif is conserved and has weak affinity to Clathrin DPF (Aspartic acid-Proline-Phenylalanine) motifs. NPF (Asparagine-Proline-Phenylalanine) motif binds to AP2 and EH domains (Eps15 Homology domain). Four phosphorylation sites (S16, Y96, S107 and S303) and two ubiquitination sites (K318 and K324) have been described in more than 5 references and validated by mass-spectrometry and thus are highlighted [65]. Altogether, the reported PTMs of PICALM are 41 phosphorylation (S2, S5, T11, S16, T18, S20, S23, T82, Y88, Y96, S107, Y138, T146, T158, Y237, S297, T301, S303, S307, S308, T312, S315, S353, T355, T356, S359, T363, T370, T379, S381, S444, S450, S453, T472, S474, K515, K534, S537, S543, T573 and S645), 15 ubiquitination (K24, K112, K134, K163, K231, K252, K288, K291, K318, K324, K336, K515, K559, K570 and K571), 1 acetylation (K28), 1 mono-methylation (R9), 1 di-methylation (R636) and 1 sumoylation (K238). (B) Computational modeling of the structure of PICALM N-terminal ANTH domain is shown (https://www.rcsb.org/structure/3ZYM (accessed on 1 June 2022)) [66]. PICALM ANTH domain is constituted of 11 α-helices. Three known phosphorylation sites (S16, Y96 and S107) in ANTH domain are highlighted in pink.