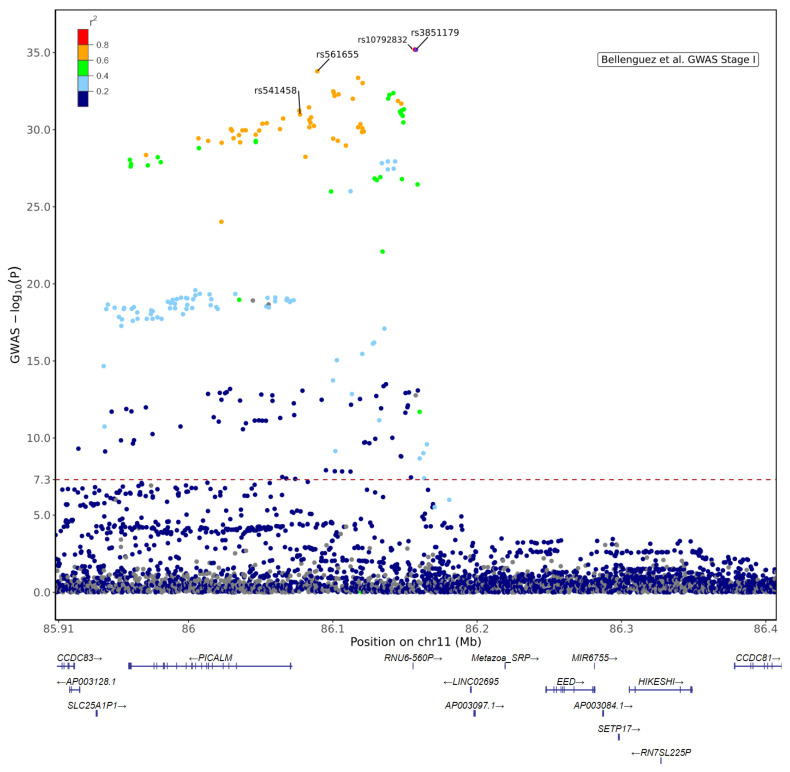
Figure 5.
Regional plot for PICALM locus based on Stage I results from [17] (Accession No GCST90027158, https://www.ebi.ac.uk/gwas/Study (accessed on 1 July 2022)). The plot shows the lead variant in Stage I + II meta-analysis, rs3851179 and +/− 250 kb extended coordinates for n = 3988 variants in chr11:85,907,598–86,407,598 (GRCh38). LD reference data derived from 1000 Genomes Project phased biallelic SNV and INDEL autosomal genetic variants called de novo GRCh38 (for selected n = 404 unrelated Non-Finnish European (NFE) sample) [85]. The lead variant rs3851179 is located in the intergenic region between PICALM and EED in which there are other non-coding genes (RNU6-560P, LINC02695 and Metazoa SRP) and a contigs (AP003097). rs10792832 (red) is in almost full LD with rs3851179. Other genome-wide significant SNPs, such as rs561655 [82,86] and rs541458 [15,87], are also in high LD with rs3851179. The red dashed line indicates the genome-wide significance threshold (p = 5 × 10−8).