Abstract
Varicella-zoster virus (VZV) is the etiological agent of varicella (chickenpox) and herpes zoster (shingles). VZV infections are ubiquitous and highly contagious, and diagnosis is mostly based on the assessment of signs and symptoms. However, monkeypox, an emerging infectious disease caused by the monkeypox virus (MPXV), has clinical manifestations that are similar to those of VZV infections. With the recent monkeypox outbreak in non-endemic regions, VZV infections are likely to be misdiagnosed in the absence of laboratory testing. Considering the lack of accessible diagnostic tests that discriminate VZV from MPXV or other poxviruses, a handy and affordable detection system for VZV is crucial for rapid differential diagnosis. Here, we developed a new detection method for VZV using recombinase-aided amplification technology, combined with the lateral flow system (RAA-LF). Given the prevalence of VZV worldwide, this method can be applied not only to distinguish VZV from other viruses causing rash, but also to foster early detection, contributing substantially to disease control.
Keywords: varicella, chickenpox, monkeypox, smallpox, RAA, lateral flow, diagnostics, point-of-care-test
1. Introduction
Varicella-zoster virus (VZV), also known as human herpesvirus 3 (HHV-3), is an α-herpesvirus that is the etiological agent of varicella (chickenpox) as a primary infection and herpes zoster (shingles) as a recurring infection [1,2,3,4,5]. VZV is specific to humans [4,6], and is extremely contagious via airborne transmission [3,4,6]. Most VZV infections result in varicella, causing fever and a generalized pruritic rash, which usually occurs during childhood [1,3,6]. Sharing a common feature of herpesviruses, VZV undergoes latency and may reactivate. Generally occurring in adults, VZV reactivation causes herpes zoster (HZ), producing a localized, painful rash [1,2,4,6]. Both varicella and zoster are prevalent worldwide [1,3], and can be serious, especially in children and adults with weakened immune systems [1,4,6].
Varicella is a rather neglected illness that has not been given much attention. To some extent, it is considered more of a nuisance than an actual disease [5,7]. Although VZV does not belong to the family of poxviruses, varicella is commonly called chickenpox, likely due to its resemblance to variola (smallpox) virus infections [8], albeit with milder symptoms [7]. Smallpox has been declared eradicated since 1980 by the World Health Organization, but other orthopoxviruses still pose a substantial threat to public health [9]. Monkeypox virus (MPXV), the next-of-kin to variola virus (VARV), has emerged as the most important orthopoxvirus in the post-smallpox era [8,10,11,12,13]. MPXV is the causative agent of monkeypox, a rare zoonosis endemic to West and Central Africa [8,10,14]. However, since early May 2022, an outbreak of monkeypox has been ongoing in non-endemic regions, such as Europe and the Americas [15,16].
The clinical manifestations of varicella are nearly identical to those of monkeypox, making them generally indistinguishable without laboratory testing [11,12,17,18]. Varicella and monkeypox coinfection has also previously been reported [19,20]. Hence, confusion in the diagnosis of varicella and monkeypox is common in areas where the viruses coexist, particularly in Central Africa [8,12,18]. Laboratory diagnostics are essential for the identification and surveillance of these diseases [11,18], and there are several laboratory tests, but they are not always available due to limited diagnostic resources in developing regions [2,3,5]. The “gold standard” for the diagnosis of VZV infections was once virus isolation, but this is time-consuming, costly, and not readily accessible [2,8,15]. This method has now been replaced by polymerase chain reaction (PCR) and direct fluorescence assays (DFA) as the methods of choice [1,11,12]. PCR, including quantitative PCR (qPCR), is considered the most sensitive and reliable testing method for VZV to date [1,11]. However, it requires expensive reagents and equipment, as well as a high degree of skill and laboratory experience [3,11].
Cases of human monkeypox are more common in Africa, where surveillance can be more challenging due to poor infrastructure and lack of healthcare facilities [11,21]. The Center for Disease Control and Prevention (CDC) has recommended a number of laboratory procedures to confirm monkeypox infections. However, these procedures are mostly based on nucleic acid amplification testing (NAAT) via PCR and/or sequencing [22,23,24,25,26,27,28]. Before its emergence in non-endemic regions in 2022, monkeypox was a neglected tropical illness that most people were not even aware of. Thus, research to develop more efficient detection methods for MPXV has lagged until recently. Development of detection tests (e.g., loop-mediated isothermal amplification, LAMP; recombinase polymerase amplification, RPA; restriction length fragment polymorphism, RLFP) for MPXV is becoming a subject of active research due to the recent outbreak [29,30,31], but that is not the case for VZV. However, since clinical distinction among varicella, monkeypox, and other rash illnesses is almost impossible in the absence of a diagnostic test, new tests are needed for a more precise and rapid diagnosis [11,18].
Here, we developed a rapid detection method for VZV using recombinase-aided amplification-lateral flow (RAA-LF) technology. The gene of interest is the VZV open reading frame 63 (ORF63), which encodes an immediate early protein (IE63) that is synthesized during lytic infection [32]. IE63 is also expressed during the latent phase, indicating a critical role in the maintenance and establishment of virus latency [33,34,35]. The whole process, from the RAA reaction to the visualization of the band on the LF dipstick, can be completed in less than 30 min.
2. Materials and Methods
2.1. Sample Description and DNA Extraction
The samples used in this study were collected during investigations carried out by the Institut Pasteur of Bangui (IPB) in the Central African Republic (CAR), when suspected cases of MPXV was reported by the health authorities. On each occasion, IPB conducted field missions to collect biological samples (crust, serum, or pus) and epidemiological information for each reported case. From the DNA extracted from these samples, IPB investigated the etiological cause by determining whether the agent was MPXV or VZV. When a suspected case of MPXV is reported, investigation for both MPXV and VZV is done systematically by IPB. The case is confirmed to be either MPXV or VZV, or occasionally, a co-infection of both viruses. Of the 20 samples, 15 were collected from crusts of lesions, and the other five were from pus. Detailed information on the samples is reported in Table 1.
Table 1.
Details of the collected varicella-zoster virus (VZV) samples.
| Sample | Year | Location | Type | Age 1 | Gender 2 |
|---|---|---|---|---|---|
| 1 | 2018 | Rafaï | crust | 29 y.o. | M |
| 2 | Rafaï | crust | 8 m.o. | M | |
| 3 | Bangui | crust | 34 y.o. | M | |
| 4 | Bangui | pus | 13 y.o. | M | |
| 5 | Bangassou | crust | unspecified | M | |
| 6 | Bangassou | crust | unspecified | unspecified | |
| 7 | Bangassou | crust | unspecified | F | |
| 8 | Bangassou | crust | unspecified | F | |
| 9 | Bangui | pus | 41 y.o. | M | |
| 10 | 2019 | Bimbo | crust | 45 y.o. | M |
| 11 | unspecified | crust | 21 y.o. | F | |
| 12 | Bangui | crust | 3 m.o. | M | |
| 13 | Mbaïki | pus | 28 y.o. | M | |
| 14 | Bria | pus | 36 y.o. | M | |
| 15 | 2020 | Boda | crust | 11 y.o. | F |
| 16 | Mbaïki | pus | 27 y.o. | M | |
| 17 | Nola | crust | 45 y.o. | M | |
| 18 | 2021 | Bimbo | crust | 5 y.o. | F |
| 19 | Bimbo | crust | 5 y.o. | M | |
| 20 | Bimbo | crust | 10 y.o. | M |
1 Age: y.o.—years old; m.o.—months old; 2 Gender: M—male; F—female.
The phenol-chloroform method was employed to isolate nucleic acid as described below. One milliliter of the extraction solution (ES) consisting of 100 mM EDTA (Invitrogen, Waltham, MA, USA), 200 mM NaCl (Invitrogen, Waltham, MA, USA), 50 mM Tris-HCI (pH 8.0) (Solarbio Life Sciences, Beijing, China), 0.5% SDS (Solarbio Life Sciences, Beijing, China), and 50 μg/mL RNase (Sigma-Aldrich, Burlington, MA, USA) was added to the sample, followed by the addition of 20 μL Proteinase K (100 μg/mL) (TIANGEN Biotech Co., Ltd., Beijing, China) and gentle mixing. The solution was incubated at 55 °C for at least 2 h with occasional mixing. An equal volume of phenol (Solarbio Life Sciences, Beijing, China) was added to the solution, and was vortexed vigorously for 1 min. The tube was centrifuged at 9600× g for 5 min at 4 °C to separate the two phases. The aqueous (top) phase was then transferred to a new tube, and an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) (Solarbio Life Sciences, Beijing, China; Sinopharm Chemical Reagent Co., Ltd., Shanghai, China; Richjoint Chemical Reagents Co., Ltd., Shanghai, China) was added to the aqueous phase. The tube was vortexed for about 1 min, and was centrifuged at 12,000 rpm for 5 min at 4 °C to separate the two phases. The aqueous phase was again transferred to a new tube, and an equal volume of chloroform:isoamyl alcohol (24:1) (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China; Richjoint Chemical Reagents Co., Ltd., Shanghai, China), was added to the aqueous phase. The tube was mixed, and was centrifuged at 9600× g for 5 min at 4 °C to separate the two phases. The aqueous phase was transferred again to a new tube. Then, 14 µL of 5M NaCl with a final concentration of 0.14 mol/l was added to the mixture, and the solution was mixed thoroughly. Two volumes of absolute ethanol (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) were added to the aqueous phase, and the solution was mixed gently. The tube was kept at −20 °C for 30 min, and was centrifuged at 9600× g for 5 min. The DNA pellet was washed with 70% ethanol twice to decrease the residual salt, and was air-dried at 37 °C to let the ethanol evaporate. The ethanol-free DNA was then dissolved in 50 μL nuclease-free water (Invitrogen, Waltham, MA, USA).
2.2. Primer and Probe Design
Primers and probes targeting the ORF63 gene of VZV (GenBank: NC_001348.1:110581-111431) were manually selected for qPCR and RAA-LF assays (Table 2). RAA primers and probes were designed in accordance with the TwistAmp® nfo kit assay design manual guidelines (www.twistdx.co.uk) (accessed on 26 July 2022). To assess the feasibility of amplification of the RAA primers, conventional PCR was performed in duplicate. The 50 µL PCR reaction consisting of 25 µL DreamTaq™ Hot Start Green PCR Master Mix (Thermo Scientific™, Waltham, MA, USA), 1 µL forward primer (10 µM), 1 µL reverse primer (10 µM), 5 µL DNA template, and 18 µL nuclease-free water was run using the MiniAmp Thermal Cycler (Applied Biosystems™, Waltham, MA, USA) under the following conditions: initial denaturation at 95 °C for 1 min, 25 cycles of 95 °C for 30 s (denaturation), 62 °C for 30 s (annealing), and 72 °C for 1 min (extension), and final extension at 72 °C for 5 min. Amplification was verified by agarose gel electrophoresis, and the gel was visualized under UV light using the Tanon 4200SF gel imaging system (Tanon Science & Technology Co., Ltd., Shanghai, China). The corresponding sequences of all primers and probes (Sangon Biotech Co., Ltd., Shanghai, China) used in this study are listed in Table 2.
Table 2.
Primers and probes to detect VZV using quantitative polymerase chain reaction (qPCR) and recombinase-aided amplification-lateral flow (RAA-LF) assays.
| Name | Sequence (5′–3′) |
|---|---|
| qPCR F | CGCGTTTTGTACTCCGGG |
| qPCR R | CGGTTGATGTCCTCAACGAG |
| qPCR P | FAM-TGGGAGATCCACCCGGCCAG-TAMRA |
| RAA-LF F1 | GATGTTAACGGAAAGATGGAATATGGATCTGC |
| RAA-LF R1 | Biotin-CGACCCATTAGATAAAAGTCGAGGCATATG |
| RAA-LF P1 | FAM-GTACTCCGGGTTGGGAGATCCACCCGGCCAGGCTC /idSp/GTTGAGGACATCAACCG-C3Sp |
2.3. RealTime-PCR (qPCR)
The VZV qPCR detection was designed to determine the concentration of the VZV plasmid in different dilutions, as well as to quantify the viral load of the VZV samples. A total volume of 20 µL qPCR reaction was comprised of 10 µL NovoStart® Probe qPCR SuperMix (Novoprotein Scientific, Inc., Suzhou, China), 1 µL qPCR forward primer (10 µM), 1 µL reverse primer (10 µM), 0.4 µL probe (10 µM), 6.6 µL nuclease-free water, and 1 µL DNA template. qPCR cycling parameters were set to 95 °C for 5 min, plus 40 cycles of 95 °C for 15 s and 60 °C for 1 min, and was run using the Applied Biosystems QuantStudio 1 Real-Time PCR System (Applied Biosystems™, Waltham, MA, USA).
2.4. Recombinase-Aided Amplification–Lateral Flow
To visually detect VZV on a dipstick, the RAA-LF assay was conducted using the RAA nfo kit ZC Bio-Sci & Tech Co., Ltd., Hangzhou, China) and the HybriDetect universal lateral flow assay kit (Milenia Biotec GmbH, Geissen, Germany). The RAA nfo reaction consisting of 25 µL Buffer A, 2 µL forward primer (2 µM), 2 µL biotinylated reverse primer (2 µM), 0.6 µL probe (2 µM), and 15.9 µL nuclease-free water, was added to one tube of RAA lyophilized powder. The reaction was mixed thoroughly, followed by the addition of 2 µL DNA (in the tube), and 2.5 µL Buffer B (on the lid), respectively. The resulting 50 µL reaction was mixed vigorously before incubating at 37 °C for 10 min in MiniAmp Thermal Cycler (Applied Biosystems™, Waltham, MA, USA). For the LF dipstick assay, 2 µL of the amplification product was diluted with 98 µL of HybriDetect Assay Buffer. For each sample to be analyzed, 100 µL of the same buffer was prepared in another tube. Ten microliters of the diluted amplified product was pipetted on to the sample application area of the dipstick, and the dipstick was immediately placed into the solution in an upright position for no more than 10 min. Amplification was confirmed by the appearance of the test band along with the control band on the dipstick.
2.5. Assay Sensitivity and Specificity
The sensitivity of the qPCR and RAA-LF methods was assessed using a recombinant puc57 plasmid with the ORF63 gene in full length (Sangon Biotech Co., Ltd., Shanghai, China) as the standard. The concentration of the undiluted plasmid was quantified using the Qubit 4 fluorometer (Invitrogen, Waltham, MA, USA). The value obtained was needed to calculate the corresponding copy number using the following equation:
| Number of copies = [plasmid concentration (ng/µL) × (6.0221 × 1023 molecules/mole)] ÷ [total plasmid length (bp) × 660 g/mole × (1 × 109 ng/g)]; where, total plasmid length = vector length (bp) + fragment length (bp) |
The limit of detection (LOD) was verified by qPCR, in triplicate, using several dilutions of VZV plasmid solutions with known copy numbers. A standard curve was established using these dilutions. The LOD determination by RAA-LF was also conducted using the same plasmid dilutions. The specificity of the RAA-LF assay was determined using VZV, MPXV, and vaccinia virus (VACV) DNA as templates. The viral DNA of MPXV was provided by IPB. A culture of VACV (ATCC® VR-2047™) was obtained from the Pathogen Discovery and Big Data Platform of the Institut Pasteur of Shanghai–Chinese Academy of Sciences (IPS-CAS). VACV was then cultivated in Vero cells prior to DNA extraction using the GeneJET Viral DNA and RNA Purification kit (Thermo Scientific™, Waltham, MA, USA) according to the manufacturer’s instructions.
3. Results
3.1. Sampling Information
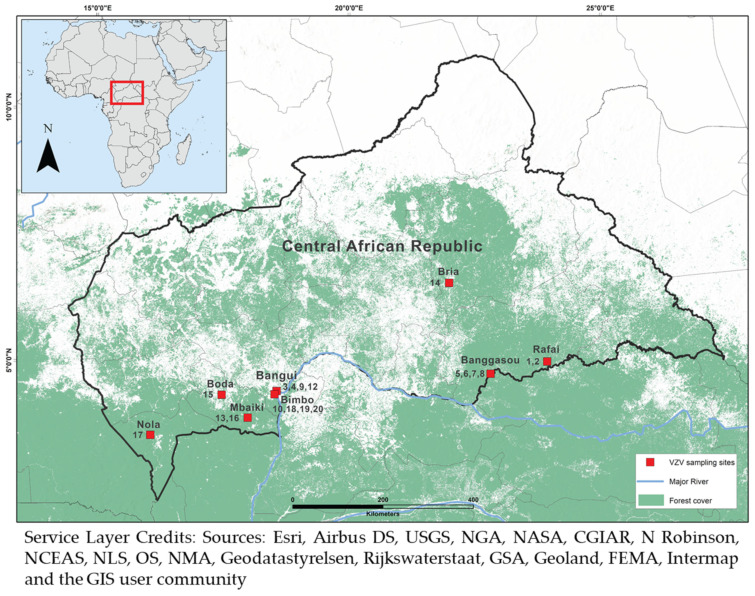
A total of 20 samples was collected over a four-year period (2018–2021) from different cities in the CAR, including Rafaï, Bangassou, Mbaïki, Bria, Boda, Nola, Bimbo, and the capital city Bangui. The majority (14) of the samples was acquired from male patients, with five from female patients, and one of unspecified gender (Table 1). With respect to age, nine samples were obtained from adults over 18 years of age, seven from children under 18, and four from patients of unknown age (Table 1). Sampling sites were mapped using the ArcGIS® software (Esri, Redlands, CA, USA) (Figure 1).
Figure 1.
Sampling sites in the Central African Republic (CAR). Twenty VZV samples were collected from various locations in the CAR from 2018 to 2021.
3.2. Primer Validation
Four sets of primers were manually designed for RAA-LF. The specificity of all primers and probes were confirmed using the NCBI BLAST tool to ensure the absence of cross-reactivity with related viral sequences. To validate the amplification feasibility of the primer pairs, conventional PCR assay and agarose gel electrophoresis were conducted. Results show that amplification was successful for all primer pairs, but a clearer band on the gel was produced by the first set of primers with an amplicon size of 227 bp (Figure S1). Primer set 1 was thus the primer pair of choice for the subsequent steps, and was used to derive the corresponding nfo probe. For the RAA nfo assay, the 5′ end of the chosen reverse primer (RAA-LF R1) was biotinylated (Table 2).
3.3. Assay Sensitivity and Specificity
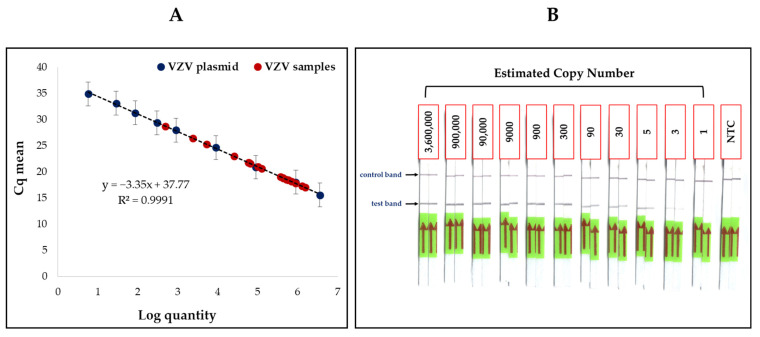
A recombinant plasmid containing the full-length ORF63 gene of VZV with a copy number of approximately 3.60 × 109 copies/µL was used to determine the detection threshold for of qPCR and RAA-LF. Plasmid dilutions with copy numbers ranging from 3.60 × 106 to 5 copies/µL were initially used to determine the LOD by qPCR. A standard curve was also plotted using these dilutions (Figure 2A, dark blue dots), and was used to determine the copy number of the VZV samples. The curve gave an R2 value of 0.9991 and a slope (m) value of −3.35, suggesting a nearly linear trend and high PCR efficiency. The LOD for VZV was also determined using RAA-LF, in duplicate, with the same dilutions mentioned above for qPCR (Figure 2B). However, since intense bands were still observed even at a low concentration of 5 copies/µL, the RAA-LF assay was performed for two additional dilutions with an approximate copy number of 3 and 1 copies/µL, respectively. Slight test bands were observed in samples with 3 copies/µL, but was absent at 1 copies/µL concentration (Figure 2B). Figure 2 shows that both qPCR and RAA-LF were able to detect VZV at a very low concentration using the standard plasmid.
Figure 2.
Limit of detection for VZV. qPCR and RAA-LF were conducted to determine the detection threshold for VZV with a recombinant plasmid. A standard curve was established using the mean threshold (Cq) values of the plasmid dilutions ((A), dark blue dots) obtained by qPCR, showing detection of VZV in samples with as little as 5 copies/µL. The mean Cq values of the VZV samples ((A), red dots) were also plotted on the standard curve against the VZV plasmid, showing a nearly linear regression line. The RAA-LF assay exhibited results similar to that of qPCR (B), with the test band present in samples with an estimated copy number of 5 copies/µL, and the no-template control (NTC) displaying only the control band. Both qPCR and RAA-LF are thus highly sensitive methods in detecting VZV.
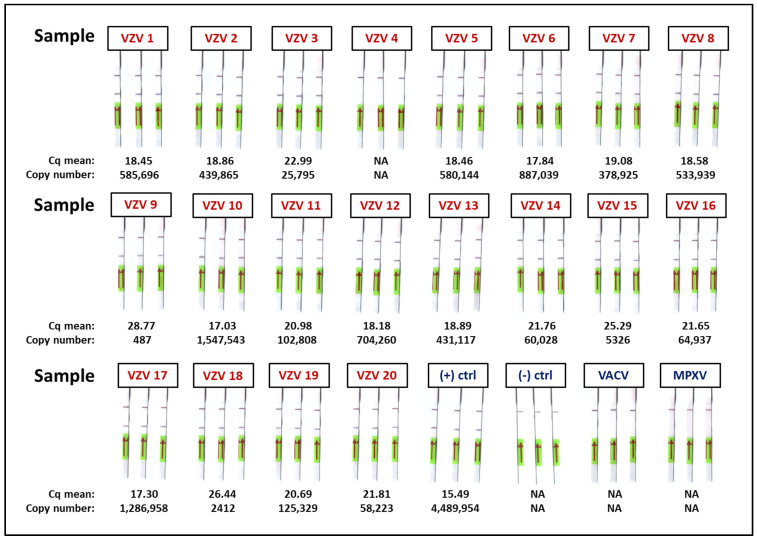
The specificity of RAA-LF was also verified using MPXV and VACV DNA as templates. The test was conducted in triplicate, and the viruses tested negative for VZV, suggesting that this test is highly specific to VZV (Figure 3).
Figure 3.
Detection of VZV in clinical samples. Twenty VZV samples were tested with qPCR, but one (sample 4) tested negative for VZV. The positive control used was the VZV plasmid with a concentration of about 3.62 × 106 copies/µL, and nuclease-free water for the negative control. The specificity of RAA-LF was determined using the DNA of monkeypox virus (MPXV) and vaccinia virus (VACV), and both viruses tested negative for VZV. RAA-LF for VZV detection was also conducted, in triplicate, using the same clinical samples tested with qPCR. Nineteen samples, along with the positive control, tested positive for VZV by RAA-LF with the presence of the test band on the dipstick, while sample 4 only exhibited the control band, conforming to the results of the qPCR.
3.4. VZV Detection in Clinical Samples with qPCR and RAA-LF
qPCR was performed in triplicate to confirm the presence of VZV in 20 samples. Nineteen of the 20 samples tested positive for VZV by qPCR, excluding sample 4, which tested negative. Sample 4 was collected from a patient with pustules that tested positive for VZV at IPB, but by the time this experiment was conducted at IPS-CAS, the sample was degraded. This negative result was verified after running another qPCR in duplicate. The corresponding mean threshold (Cq) value and copy number for each sample are presented in Figure 3. These results show that VZV was detected with copy numbers ranging from 487 copies/µL to 1.55 × 106 copies/µL (samples 9 and 10, respectively). RAA-LF was also conducted, in triplicate, for the 20 clinical samples tested with qPCR. The presence of both the test band and the control band was observed, except for sample 4, which showed only the control band, in accordance with the qPCR results. The summary of the results is presented in Table 3.
Table 3.
Results of RT-PCR and RAA-LF tests using clinical samples.
| Method | Positive | Negative | Total | PPV 1 | NPV 2 |
|---|---|---|---|---|---|
| RT-PCR | 19 | 1 | 20 | 1 | 1 |
| RAA-LF | 19 | 1 | 20 | 1 | 1 |
1 PPV—positive predictive value; 2 NPV—negative predictive value.
4. Discussion
VZV is ubiquitous in nature, and laboratory confirmation is not routinely practiced to detect VZV infections. The samples used in this study were collected from the skin lesions of patients initially suspected to be infected with monkeypox, but turned out to be infected with VZV. Due to concurrent cases of VZV and MPXV, diagnosis based on clinical grounds alone is challenging and laboratory testing is necessary. The cases of VZV infections in the CAR were detected in the same cities and towns where monkeypox cases were also observed. These localities are close to remote forest areas where spillover events, including the enzootic monkeypox, occur [36,37,38] (Figure 1). Since outbreaks of VZV and MPXV infections have been reported simultaneously in the same regions, it is imperative to develop a rapid detection method not only for VZV, but also for MPXV, and potentially other poxviruses as well. With the current emergence of monkeypox as an infectious disease, proper clinical response for rash patients relies on a rapid detection test readily available to distinguish between VZV and MPXV during outbreaks.
Given its rarity, MPXV can be mistaken for VZV if it is diagnosed based entirely on clinical presentation, especially in remote areas where laboratory testing cannot be performed. Considering that diagnostic tools for both VZV and MPXV are limited, a rapid detection system for both viruses is necessary in case such situations arise. Isothermal nucleic acid amplification tests (iNAT), such as LAMP and RPA, have long been established for pathogen detection, and have been reported as good PCR alternatives [29,31]. However, one drawback of the LAMP assay is that it requires multiple sets of primers, in contrast to RAA/RPA, in which only a single pair of primers and a probe are necessary. A highly specific molecular detection system for MPXV has also been developed by our team using recombinase-based isothermal amplification techniques. Although orthopoxviruses are highly cross-reactive, three different RAA/RPA approaches (including visualization of the amplified product on a dipstick) have proven greatly successful in detecting MPXV in clinical samples [39].
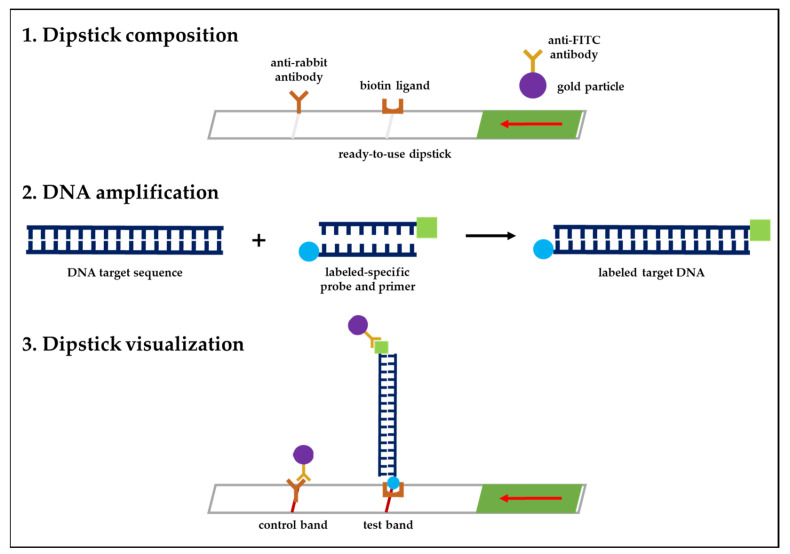
The RAA technique combined with the LF system was employed in this study to develop a detection method for VZV. As an enzymatic procedure, RAA is a brilliant scientific breakthrough that has already been successfully integrated in different detection strategies [40]. HybriDetect, on the other hand, is a ready-to-use test strip based on lateral flow technology using gold particles. The dipstick was designed to rapidly detect analytes, such as gene amplicons, labeled with FITC (probe) and biotin (primer). In this study, an analyte solution containing a specific probe labeled with FITC and a primer labeled with biotin was developed, and the sample was mixed with the analyte solution where the test strip was placed. The labeled analyte binds to the gold-labeled FITC-specific antibodies in the sample application area of the strip. Driven by capillary forces, travel through the membrane. On the one hand, only analyte-captured gold particles bound to immobilized biotin-ligand molecules as they passed through the line, generating a red-blue band over time. On the other hand, unbound gold particles migrated over the control band, where they were captured by species-specific antibodies. The mechanism of the HybriDetect universal lateral flow dipstick is presented Figure 4 (https://www.milenia-biotec.com/en/product/hybridetect/#nav-protocol) (accessed on 26 October 2022). For the detection method we developed for VZV—and as in any other tests that make use of the LF system for pathogen detection—the occurrence of a false-positive line on the dipstick is inevitable. Several factors contribute to these false positives, such as false amplification due to contamination, as well as the use of incompatible primers and/or probes that can result in unexpected false-positive signals [41,42,43,44,45,46]. As in this study, we recommend incubating the dipstick in the buffer solution for no longer than 5 min to avoid false-positive results. Incubation for times longer than 5 min may trigger the appearance of a false-positive line on the dipstick.
Figure 4.
The lateral flow mechanism of the HybriDetect dipstick. (1) The sample application area of the dipstick is composed of gold-labeled FITC-specific antibodies. (2) Amplification of DNA by RAA nfo is performed using analyte detectors labeled with FITC (probe) and biotin (primer) that attach to the DNA target sequence. (3) The labeled analytes bind to the gold-labeled FITC-specific antibodies, travel to the membrane, and bind to immobilized biotin-ligand molecules while passing the test line, eventually generating a red-blue band. Unbound gold particles then migrate to the control line.
VZV is characterized by neurotropism and lifelong latent infection [47,48,49]. After primary infection, the virus gains access to neurons, establishes latency in the ganglia of the peripheral nervous system (PNS), and may later on reactivate and cause shingles [47,48,49,50,51]. The gene of interest for the RAA-LF system is the ORF63, which synthesizes a protein (IE63) that is expressed during both lytic and latent phases of infection [32]. The gene-encoded protein IE63 is located in the nucleus during lytic replication, and localizes to the cytoplasm of neurons during latency. However, in case of reactivation, the protein is present in both the nucleus and cytoplasm [52]. Although ORF63 is the most abundantly expressed gene at the levels of both protein and mRNA during latency [53,54], this RAA-LF system cannot be used to detect the latent form of VZV. Latent VZV infections are predominantly, if not exclusively, contained in the ganglia of the PNS [50,51,52], but the samples used for testing in this study are from skin lesions. Due to the mechanism of VZV latency, a positive test strip result from skin samples (not containing ganglia) would only be possible in the case of an active infection, either as a primary infection (chickenpox) or reactivation of the virus from latency as shingles [49,55].
The detection method developed for VZV in this study can be used as a point-of-care-test (POCT), especially in rural areas, because it can be easily done out of the laboratory by non-specialists. PCR may be the “gold standard” for laboratory diagnostics today, but RAA can be a useful complement and a potential substitute to this method, given its convenience and accessibility. In any case, timely detection of the causative agent is essential for infection control. Thus, having an easy, cheap, and handy detection test is advantageous in curbing possible VZV and/or MPXV outbreaks.
5. Conclusions
There is a constant need for laboratory diagnostics to determine the causative agent of a disease, and to be able to execute appropriate actions in a timely and fruitful manner. Making an accurate diagnosis for VZV infections can be difficult based on clinical manifestations alone, and, due to high VZV transmissibility, there is a vast need to develop a rapid detection method for this virus. Laboratory testing is important for detecting VZV, and PCR is the preferred diagnostic test given its accuracy and sensitivity. However, it is time-consuming and costly, and during outbreaks, timeliness is crucial. Therefore, the successful development of a convenient detection method for VZV greatly benefits the less developed areas of the world, in particular, as it saves both time and resources. In addition, it is also a potentially commercially viable product, thereby conferring great economic importance. Finally, given their convenience, rapid detection systems using RAA are likely to become the standard diagnostic method, ultimately replacing PCR, especially in regions where laboratory testing is not readily available.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/diagnostics12122957/s1, Figure S1: RAA primer screening by PCR.
Author Contributions
N.B. and G.W. conceived and supervised the project, and acquired funding. B.S., E.G. and E.N. collected the VZV samples, and performed DNA extraction from samples. K.M.B. and L.M. designed the primers and probes for RAA. K.M.B. performed the experiments. K.M.B., G.W. and N.B. analyzed the experimental results. K.M.B. wrote the first draft of the manuscript. N.B. and G.W. participated in the critical reading and editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The sample collection from confirmed VZV patients in the CAR was approved by the ethics committee of the Institut Pasteur of Bangui (authorization number UB/FACSS/IPB/CES/20) on 21 February 2021.
Informed Consent Statement
Written informed consent was obtained from the patients to publish this paper.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This project was supported by the Ministry of Science and Technology of the People’s Republic of China (Grant Nos. 2021YFC0863400, 2022YFE0114700), the Alliance of International Scientific Organizations (Grant No. ANSO-CR-SP-2020-02), the Shanghai Municipal Science and Technology Major Project (Grant No. 2019SHZDZX02), G4 funding from the Institut Pasteur, Fondation Merieux, and Chinese Academy of Sciences to G.W., and the International Affairs Department of the Institut Pasteur of Paris. This project was also supported by the French National Research Agency (Grant AFRIPOX). The funders had no role in study design, data analysis, or the preparation of the manuscript.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arvin A.M. Varicella-zoster virus. Clin. Microbiol. Rev. 1996;9:361–381. doi: 10.1128/CMR.9.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy P.G.E., Gershon A.A. Clinical Features of Varicella-Zoster Virus Infection. Viruses. 2018;10:609. doi: 10.3390/v10110609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauerbrei A. Diagnosis, antiviral therapy, and prophylaxis of varicella-zoster virus infections. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:723–734. doi: 10.1007/s10096-016-2605-0. [DOI] [PubMed] [Google Scholar]
- 4.Andrei G., Snoeck R. Advances and Perspectives in the Management of Varicella-Zoster Virus Infections. Molecules. 2021;26:1132. doi: 10.3390/molecules26041132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gershon A.A., Breuer J., Cohen J.I., Cohrs R.J., Gershon M.D., Gilden D., Grose C., Hambleton S., Kennedy P.G., Oxman M.N., et al. Varicella zoster virus infection. Nat. Rev. Dis. Prim. 2015;1:15016. doi: 10.1038/nrdp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varicella and herpes zoster vaccines: WHO position paper, June 2014—Recommendations. Vaccine. 2016;34:198–199. doi: 10.1016/j.vaccine.2014.07.068. [DOI] [PubMed] [Google Scholar]
- 7.Gershon A.A., Gershon M.D. Pathogenesis and current approaches to control of varicella-zoster virus infections. Clin. Microbiol. Rev. 2013;26:728–743. doi: 10.1128/CMR.00052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jezek Z., Szczeniowski M., Paluku K.M., Mutombo M., Grab B. Human monkeypox: Confusion with chickenpox. Acta Trop. 1988;45:297–307. [PubMed] [Google Scholar]
- 9.Berthet N., Descorps-Declère S., Besombes C., Curaudeau M., Meyong A.A.N., Selekon B., Labouba I., Gonofio E.C., Ouilibona R.S., Tchetgna H.D.S., et al. Genomic history of human monkey pox infections in the Central African Republic between 2001 and 2018. Sci. Rep. 2021;11:13085. doi: 10.1038/s41598-021-92315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore M.J., Rathish B., Zahra F. StatPearls. StatPearls Publishing; Treasure Island, FL, USA: 2022. Monkeypox. [PubMed] [Google Scholar]
- 11.McCollum A.M., Damon I.K. Human monkeypox. Clin. Infect. Dis. 2014;58:260–267. doi: 10.1093/cid/cit703. [DOI] [PubMed] [Google Scholar]
- 12.MacNeil A., Reynolds M.G., Carroll D.S., Karem K., Braden Z., Lash R., Moundeli A., Mombouli J.V., Jumaan A.O., Schmid D.S., et al. Monkeypox or varicella? Lessons from a rash outbreak investigation in the Republic of the Congo. Am. J. Trop. Med. Hyg. 2009;80:503–507. doi: 10.4269/ajtmh.2009.80.503. [DOI] [PubMed] [Google Scholar]
- 13.Petersen E., Abubakar I., Ihekweazu C., Heymann D., Ntoumi F., Blumberg L., Asogun D., Mukonka V., Lule S.A., Bates M., et al. Monkeypox-Enhancing public health preparedness for an emerging lethal human zoonotic epidemic threat in the wake of the smallpox post-eradication era. Int. J. Infect. Dis. 2019;78:78–84. doi: 10.1016/j.ijid.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yong S.E.F., Ng O.T., Ho Z.J.M., Mak T.M., Marimuthu K., Vasoo S., Yeo T.W., Ng Y.K., Cui L., Ferdous Z., et al. Imported Monkeypox, Singapore. Emerg. Infect. Dis. 2020;26:1826–1830. doi: 10.3201/eid2608.191387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarner J., Del Rio C., Malani P.N. Monkeypox in 2022-What Clinicians Need to Know. JAMA. 2022;328:139–140. doi: 10.1001/jama.2022.10802. [DOI] [PubMed] [Google Scholar]
- 16.Thornhill J.P., Barkati S., Walmsley S., Rockstroh J., Antinori A., Harrison L.B., Palich R., Nori A., Reeves I., Habibi M.S., et al. Monkeypox Virus Infection in Humans across 16 Countries—April–June 2022. N. Engl. J. Med. 2022;387:679–691. doi: 10.1056/NEJMoa2207323. [DOI] [PubMed] [Google Scholar]
- 17.Petersen E., Kantele A., Koopmans M., Asogun D., Yinka-Ogunleye A., Ihekweazu C., Zumla A. Human Monkeypox: Epidemiologic and Clinical Characteristics, Diagnosis, and Prevention. Infect. Dis. Clin. N. Am. 2019;33:1027–1043. doi: 10.1016/j.idc.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maksyutov R.A., Gavrilova E.V., Shchelkunov S.N. Species-specific differentiation of variola, monkeypox, and varicella-zoster viruses by multiplex real-time PCR assay. J. Virol. Methods. 2016;236:215–220. doi: 10.1016/j.jviromet.2016.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoff N.A., Morier D.S., Kisalu N.K., Johnston S.C., Doshi R.H., Hensley L.E., Okitolonda-Wemakoy E., Muyembe-Tamfum J.J., Lloyd-Smith J.O., Rimoin A.W. Varicella Coinfection in Patients with Active Monkeypox in the Democratic Republic of the Congo. Ecohealth. 2017;14:564–574. doi: 10.1007/s10393-017-1266-5. [DOI] [PubMed] [Google Scholar]
- 20.Hughes C.M., Liu L., Davidson W.B., Radford K.W., Wilkins K., Monroe B., Metcalfe M.G., Likafi T., Lushima R.S., Kabamba J., et al. A Tale of Two Viruses: Coinfections of Monkeypox and Varicella Zoster Virus in the Democratic Republic of Congo. Am. J. Trop. Med. Hyg. 2020;104:604–611. doi: 10.4269/ajtmh.20-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynolds M.G., McCollum A.M., Nguete B., Lushima R.S., Petersen B.W. Improving the Care and Treatment of Monkeypox Patients in Low-Resource Settings: Applying Evidence from Contemporary Biomedical and Smallpox Biodefense Research. Viruses. 2017;9:380. doi: 10.3390/v9120380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espy M.J., Cockerill F.R., III, Meyer R.F., Bowen M.D., Poland G.A., Hadfield T.L., Smith T.F. Detection of smallpox virus DNA by LightCycler PCR. J. Clin. Microbiol. 2002;40:1985–1988. doi: 10.1128/JCM.40.6.1985-1988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D., Wilkins K., Mccollum A.M., Osadebe L., Kabamba J., Nguete B., Likafi T., Balilo M.P., Lushima R.S., Malekani J., et al. Evaluation of the GeneXpert for Human Monkeypox Diagnosis. Am. J. Trop. Med. Hyg. 2017;96:405–410. doi: 10.4269/ajtmh.16-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Olson V.A., Laue T., Laker M.T., Damon I.K. Detection of monkeypox virus with real-time PCR assays. J. Clin. Virol. 2006;36:194–203. doi: 10.1016/j.jcv.2006.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Zhao H., Wilkins K., Hughes C., Damon I.K. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J. Virol. Methods. 2010;169:223–227. doi: 10.1016/j.jviromet.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson V.A., Laue T., Laker M.T., Babkin I.V., Drosten C., Shchelkunov S., Niedrig M., Damon I.K., Meyer H. Real-time PCR system for detection of orthopoxviruses and simultaneous identification of smallpox virus. J. Clin. Microbiol. 2004;42:1940–1946. doi: 10.1128/JCM.42.5.1940-1946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ropp S.L., Jin Q., Knight J.C., Massung R.F., Esposito J.J. PCR strategy for identification and differentiation of small pox and other orthopoxviruses. J. Clin. Microbiol. 1995;33:2069–2076. doi: 10.1128/jcm.33.8.2069-2076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shchelkunov S.N., Shcherbakov D.N., Maksyutov R.A., Gavrilova E.V. Species-specific identification of variola, monkeypox, cowpox, and vaccinia viruses by multiplex real-time PCR assay. J. Virol. Methods. 2011;175:163–169. doi: 10.1016/j.jviromet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davi S.D., Kissenkötter J., Faye M., Böhlken-Fascher S., Stahl-Hennig C., Faye O., Faye O., Sall A.A., Weidmann M., Ademowo O.G., et al. Recombinase polymerase amplification assay for rapid detection of Monkeypox virus. Diagn. Microbiol. Infect. Dis. 2019;95:41–45. doi: 10.1016/j.diagmicrobio.2019.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dumont C., Irenge L.M., Magazani E.K., Garin D., Muyembe J.-J.T., Bentahir M., Gala J.-L. Simple technique for in field samples collection in the cases of skin rash illness and subsequent PCR detection of orthopoxviruses and varicella zoster virus. PLoS ONE. 2014;9:e96930. doi: 10.1371/journal.pone.0096930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iizuka I., Saijo M., Shiota T., Ami Y., Suzaki Y., Nagata N., Hasegawa H., Sakai K., Fukushi S., Mizutani T., et al. Loop-mediated isothermal amplification-based diagnostic assay for monkeypox virus infections. J. Med. Virol. 2009;81:1102–1108. doi: 10.1002/jmv.21494. [DOI] [PubMed] [Google Scholar]
- 32.Lynch J.M., Kenyon T.K., Grose C., Haya J., Ruyechan W.T. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology. 2002;302:71–82. doi: 10.1006/viro.2002.1555. [DOI] [PubMed] [Google Scholar]
- 33.Cohen J.I., Cox E., Pesnicak L., Srinivas S., Krogmann T. The varicella-zoster virus open reading frame 63 latency-associated protein is critical for establishment of latency. J. Virol. 2004;78:11833–11840. doi: 10.1128/JVI.78.21.11833-11840.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller N.H., Walters M.S., Marcus R.A., Graf L.L., Prenni J., Gilden D., Silverstein S.J., Cohrs R.J. Identification of phosphorylated residues on varicella-zoster virus immediate-early protein ORF63. Pt 5J. Gen. Virol. 2010;91:1133–1137. doi: 10.1099/vir.0.019067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Debrus S., Sadzot-Delvaux C., Nikkels A.F., Piette J., Rentier B. Varicella-zoster virus gene 63 encodes an immediate-early protein that is abundantly expressed during latency. J. Virol. 1995;69:3240–3245. doi: 10.1128/jvi.69.5.3240-3245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Besombes C., Gonofio E., Konamna X., Selekon B., Grant R., Gessain A., Berthet N., Manuguerra J.-C., Fontanet A., Nakouné E. Intrafamily Transmission of Monkeypox Virus, Central African Republic, 2018. Emerg. Infect. Dis. 2019;25:1602–1604. doi: 10.3201/eid2508.190112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khodakevich L., Widy-Wirski R., Arita I., Marennikova S.S., Nakano J., Meunier D. Monkey pox virus infection in humans in the Central African Republic. Bull. Soc. Pathol. Exot. Fil. 1985;78:311–320. [PubMed] [Google Scholar]
- 38.Nakoune E., Lampaert E., Ndjapou S.G., Janssens C., Zuniga I., Van Herp M., Fongbia J.P., Koyazegbe T.D., Selekon B., Komoyo G.F., et al. A Nosocomial Outbreak of Human Monkeypox in the Central African Republic. Open Forum Infect. Dis. 2017;4:ofx168. doi: 10.1093/ofid/ofx168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao L., Ying J., Selekon B., Gonofio E., Wang X., Nakoune E., Wong G., Berthet N. Development and Characterization of Recombinase-Based Isothermal Amplification Assays (RPA/RAA) for the Rapid Detection of Monkeypox Virus. Viruses. 2022;14:2112. doi: 10.3390/v14102112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordray M.S., Richards-Kortum R.R. A paper and plastic device for the combined isothermal amplification and lateral flow detection of Plasmodium DNA. Malar. J. 2015;14:472. doi: 10.1186/s12936-015-0995-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Georgoutsou-Spyridonos M., Filippidou M., Kaprou G.D., Mastellos D.C., Chatzandroulis S., Tserepi A. Isothermal Recombinase Polymerase Amplification (RPA) of E. coli gDNA in Commercially Fabricated PCB-Based Microfluidic Platforms. Micromachines. 2021;12:1387. doi: 10.3390/mi12111387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J., Ma B., Fang J., Zhi A., Chen E., Xu Y., Yu X., Sun C., Zhang M. Recombinase Polymerase Amplification (RPA) Combined with Lateral Flow Immunoassay for Rapid Detection of Salmonella in Food. Foods. 2019;9:27. doi: 10.3390/foods9010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McQuillan J.S., Wilson M.W. Recombinase polymerase amplification for fast, selective, DNA-based detection of faecal indicator Escherichia coli. Lett. Appl. Microbiol. 2021;72:382–389. doi: 10.1111/lam.13427. [DOI] [PubMed] [Google Scholar]
- 45.Wang L., Zhao P., Si X., Li J., Dai X., Zhang K., Gao S., Dong J. Rapid and Specific Detection of Listeria monocytogenes with an Isothermal Amplification and Lateral Flow Strip Combined Method That Eliminates False-Positive Signals From Primer-Dimers. Front. Microbiol. 2019;10:2959. doi: 10.3389/fmicb.2019.02959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H., Zhao P., Yang X., Li J., Zhang J., Zhang X., Zeng Z., Dong J., Gao S., Lu C. A Recombinase Polymerase Amplification and Lateral Flow Strip Combined Method That Detects Salmonella enterica Serotype Typhimurium with No Worry of Primer-Dependent Artifacts. Front. Microbiol. 2020;11:1015. doi: 10.3389/fmicb.2020.01015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadaoka T., Depledge D.P., Rajbhandari L., Venkatesan A., Breuer J., Cohen J.I. In vitro system using human neurons demonstrates that varicella-zoster vaccine virus is impaired for reactivation, but not latency. Proc. Natl. Acad. Sci. USA. 2016;113:E2403–E2412. doi: 10.1073/pnas.1522575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tommasi C., Breuer J. The Biology of Varicella-Zoster Virus Replication in the Skin. Viruses. 2022;14:982. doi: 10.3390/v14050982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rentier B., Piette J., Baudoux L., Debrus S., Defechereux P., Merville M.-P., Sadzot-Delvaux C., Schoonbroodt S. Lessons to be learned from varicella-zoster virus. Vet. Microbiol. 1996;53:55. doi: 10.1016/S0378-1135(96)01234-5. [DOI] [PubMed] [Google Scholar]
- 50.Azarkh Y., Gilden D., Cohrs R.J. Molecular characterization of varicella zoster virus in latently infected human ganglia: Physical state and abundance of VZV DNA, Quantitation of viral transcripts and detection of VZV-specific proteins. Curr. Top. Microbiol. Immunol. 2010;342:229–241. doi: 10.1007/82_2009_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kennedy P.G., Grinfeld E., Gow J.W. Latent varicella-zoster virus is located predominantly in neurons in human trigeminal ganglia. Proc. Natl. Acad. Sci. USA. 1998;95:4658–4662. doi: 10.1073/pnas.95.8.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen J.I., Krogmann T., Bontems S., Sadzot-Delvaux C., Pesnicak L. Regions of the varicella-zoster virus open reading frame 63 latency-associated protein important for replication in vitro are also critical for efficient establishment of latency. J. Virol. 2005;79:5069–5077. doi: 10.1128/JVI.79.8.5069-5077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Depledge D.P., Sadaoka T., Ouwendijk W.J.D. Molecular Aspects of Varicella-Zoster Virus Latency. Viruses. 2018;10:349. doi: 10.3390/v10070349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kennedy P.G.E., Mogensen T.H., Cohrs R.J. Recent Issues in Varicella-Zoster Virus Latency. Viruses. 2021;13:2018. doi: 10.3390/v13102018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eshleman E., Shahzad A., Cohrs R.J. Varicella zoster virus latency. Future Virol. 2011;6:341–355. doi: 10.2217/fvl.10.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.