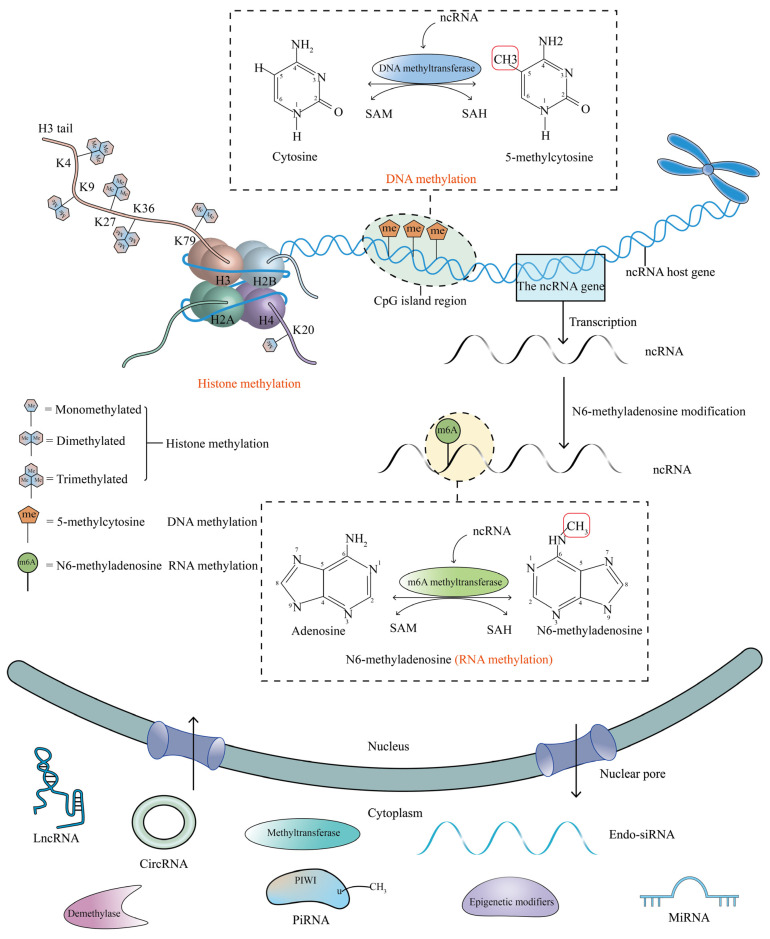
Figure 1.
DNA methylation of the ncRNA host gene, m6A modification of ncRNA, and chromatin regulation by histone modifications. DNA methylation refers to a kind of methylation modification on DNA, which is under the action of DNA methyltransferase, S-adenosyl methionine (SAM) acts as a methyl donor to covalently add a methyl group at the cytosine 5 carbon position of the CpG dinucleotide in the genome, and SAM is transformed into SAH (S-adenosylhomocysteine). N6-methyladenosine is a type of RNA methylation modification which is the m6A methyltransferase that adds a methyl group to the N6 position of adenosine, converting SAM to SAH. Histones are the core components of the nucleosome subunits in which histones H3, H4, H2A, and H2B form an octamer surrounded by DNA fragments. Histone methylation is a covalent modification that usually occurs on H3 and H4 lysine residues, which can be monomethylated, dimethylated, even trimethylated. K4, K9, K27, K36, and K79 of H3 and K20 of histone H4 are common sites of histone lysine methylation. These catalytic processes are reversible, and the red box represents the added methyl group. The ncRNAs undergoing chromatin modification, DNA methylation modification, and m6A modification, are transferred from the nucleus to the cytoplasm through the nuclear pores, and then form mature ncRNAs in the cytoplasm. The mature ncRNAs can regulate the protein expression levels of methylation modifiers in the cytoplasm and can also enter the nucleus through the nuclear pore to regulate the function of methylation modifiers.