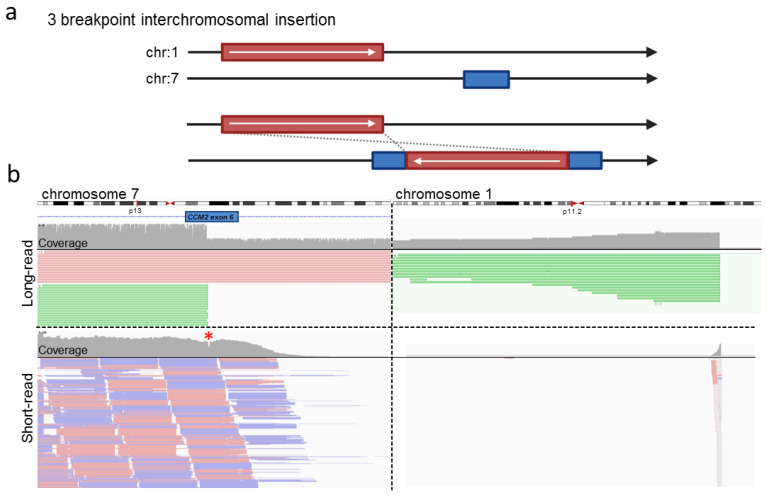
Figure 5.
Nanopore sequencing confidently detected an interchromosomal insertion in CCM2. (a) Schematic representation of the interchromosomal insertion in CCM2. (b) Long-read sequencing data of a previously described heterozygous interchromosomal insertion revealed about 50% of reads covering CCM2 terminate in exon 6 (green). These reads all had a supplementary alignment mapping to chromosome 1. Short-read data showed consistent coverage of CCM2 exon 6. The coverage at the position of the variant breakpoints was reduced (*, red star), and a limited number of reads had a supplementary alignment on chromosome 1. Most reads bridging to chromosome 1 had a mapping quality equal to 0 (hollow reads). Read data were inspected in IGV [23].